Журнал высшей нервной деятельности им. И.П. Павлова, 2022, T. 72, № 6, стр. 741-767
Подходы к применению графового анализа для исследования ЭЭГ человека в норме и при церебральной патологии
К. Д. Вигасина 1, *, Е. А. Прошина 2, П. М. Готовцев 3, 4, Е. В. Шарова 1, В. А. Бордюг 3, 4, Е. Л. Машеров 5, Г. Г. Князев 2
1 ФГБУН Институт высшей нервной деятельности и нейрофизиологии РАН
Москва, Россия
2 ФГБНУ “Научно-исследовательский институт нейронаук и медицины”
Новосибирск, Россия
3 Московский физико-технический институт
Москва, Россия
4 Национальный исследовательский центр “Курчатовский институт”
Москва, Россия
5 ФГАУ “НМИЦ нейрохирургии им. ак. Н.Н. Бурденко”
Москва, Россия
* E-mail: kristina.vigasina@yandex.ru
Поступила в редакцию 03.08.2021
После доработки 14.04.2022
Принята к публикации 26.04.2022
- EDN: OHCQZB
- DOI: 10.31857/S0044467722060107
Аннотация
Информативность распространенного и значимого для оценки функциональной активности головного мозга метода ЭЭГ существенно повышается применением математического анализа, в котором важное место занимает характеристика пространственной синхронизации, или, иными словами, функциональной коннективности биопотенциалов (на основе корреляционного и когерентного анализа, фазовой синхронизации и др.). Успехи методов нейровизуализации последних лет не только подтверждают значимость этого показателя, но и способствуют совершенствованию подходов к его статистической оценке и визуализации. К числу перспективных для анализа нейросетевой организации головного мозга методов относится графовый анализ (ГА). Его преимуществами являются наглядное описание целостной структуры сети и ее отдельных компонентов, а также определение взаимосвязей между ними. Цель настоящего обзора – на основе анализа данных литературы представить подходы к применению графового анализа и возможности данного метода. В работе представлены общие сведения о сферах применения ГА, рассматриваются наиболее распространенные и информативные метрики, приводятся рекомендации по выбору программного обеспечения. Описываются модификации ГА ЭЭГ: без первичного нахождения источников генерации составляющих ЭЭГ и с их локализацией. Приводятся примеры эффективного использования графового анализа электроэнцефалограммы здорового и больного мозга.
ВВЕДЕНИЕ
ЭЭГ – это распространенный, неинвазивный, малозатратный метод исследования функциональной активности головного мозга человека на основе регистрации и анализа биоэлектрических сигналов, отражающих совместную кооперативную деятельность большого числа функциональных единиц (Steriade et al., 1990; Гриндель, Сазонова, 2001). Паттерн энцефалограммы представляет собой суперпозицию потенциалов, генерируемых множеством пространственно разделенных источников корковой и глубинной локализации (Пономарев, 2016). Информативность энцефалографии, доказанная многолетней историей ее развития (Lopes da Silva, 2011; Шарова, 2009), существенно повышается применением методов математического анализа (Русинов, 1987; Pfurtscheller et al., 2017; Гнездицкий, 2004; Иванов, 2004; Schoffelen, Gross, 2009; Wairagkar et al., 2018; Babiloni et al., 2020). Среди них важное место занимает анализ пространственной синхронизации биопотенциалов (на основе корреляционного и когерентного анализа, фазовой синхронизации и др. методов). Согласно представлениям отечественных нейрофизиологических школ М.Н. Ливанова и В.С. Русинова (60–80-е годы прошлого века) (Ливанов, 1972; Русинов, 1969, 1987), подтвержденным последующими исследованиями (Шеповальников и др., 1997; Cвидерская, 2008; Singer, 2009; Fries, 2015; Wendelken et al., 2017), получаемые при этом показатели характеризуют уровень и особенности функциональных межцентральных связей, которые являются основой формирования вариативных состояний покоя и обеспечения разных видов активной деятельности, включая психическую. К настоящему времени выявлены наиболее общие характеристики функциональной ЭЭГ-коннективности здорового мозга и их нарушения при разных формах патологии, в том числе при угнетении сознания (Гриндель, 1980; Шарова, 2004; Boldуreva et al., 2007; Sharova et al., 2018; Cacciola et al., 2019).
В последние десятилетия мощный импульс развития получили методы нейровизуализации (фМРТ, ПЭТ, МРТ-трактография), позволяющие прижизненно, в режиме реального времени, исследовать разноуровневое (анатомия, гемодинамика, обменные процессы) структурно-функциональное обеспечение разных видов церебральной активности в норме и при патологии (Habas, 2018; Mannil, Winklhofer, 2020). Это способствовало совершенствованию методов математического анализа структурных и функциональных церебральных характеристик, а также разработке информационных теорий о нейросетевой организации деятельности мозга (Dehaene et al., 2006, 2011; Gong et al., 2009; Анохин, 2021) и функциональной коннективности как основе ее формирования (Friston et al., 1993; Мартынова и др., 2016; Храмов и др., 2021). В результате взаимного методологического обогащения развитие методов математического анализа приводит к лучшему пониманию структурных и функциональных особенностей мозга. Вместе с тем использование методов нейровизуализации имеет ряд объективных и субъективных ограничений: непереносимость процедуры некоторыми здоровыми людьми и пациентами с церебральной патологией, наличие металлических конструкций в теле, длительная вынужденная неподвижность, значительная стоимость исследования и др.
В этой связи целесообразным представляется повышение информационных возможностей ЭЭГ (как исследовательских, так и клинических) привлечением современных математических методов, хорошо зарекомендовавших себя в нейровизуализации. В частности, это относится к методам, позволяющим анализировать головной мозг как единую сеть (коннектом (Kennedy et al., 2016)) или даже гиперсеть (когнитом (Анохин, 2021)), интеграция элементов которой определяет протекание когнитивных процессов разной степени сложности. Среди таких методов значительный интерес вызывает графовый анализ (Avena-Koenigsberger et al., 2018; Girn et al., 2019), реализованный в ряде пакетов программ (toolbox) и, наряду с фМРТ (Van den Heuvel, Sporns, 2013, 2019), успешно применяемый в ряде зарубежных ЭЭГ-исследований (Chennu et al., 2014; Vecchio et al., 2017; Hatlestad-Hall et al., 2021), но недостаточно широко распространенный в России (Knyazev et al., 2015; Proshina et al., 2018).
Следует отметить, что анализ различных сложноорганизованных систем с использованием графов сегодня – это активно развивающееся направление, несмотря на то, что теория графов существует уже достаточно давно. Данный тип анализа широко применяется в электронике, сетях связи и т.д. (Larsen, 1997; Watanabe, Shinoda, 1999; Hamzeh et al., 2019). К настоящему времени подобные модели анализируют сети с сотнями узлов, что говорит о больших возможностях подхода при анализе сложных объектов и живых систем (Hamzeh et al., 2019). Таким образом, применение ГА для обработки данных ЭЭГ и фМРТ выглядит естественным расширением областей его использования. При наличии информативных публикаций, касающихся теоретического обоснования и описания возможных подходов к исследованию сетевой деятельности головного мозга (Pospelov et al., 2019; Храмов и др., 2021), многие вопросы, связанные с практическим освоением и использованием методов ГА применительно к ЭЭГ, нуждаются в дополнительном освещении.
Цель настоящей работы – на основе анализа данных литературы представить возможности метода графового анализа коннективности ЭЭГ и подходы к его применению.
Историческая справка
В 1736 году задача о семи мостах Кёнигсберга заинтересовала математика Леонарда Эйлера, члена Петербургской академии наук. В ней необходимо было определить, возможно ли пройти по всем мостам через реку Преголя, не проходя ни по одному из них дважды. Л. Эйлер доказал, что это невозможно, составив упрощенную схему города – граф, в которой мостам соответствовали линии (ребра графа), а частям города – точки соединения линий (вершины графа).
Хотя в основоположниках теории графов числят именно Л. Эйлера, однако как раздел математики она оформилась позже. Сам термин “граф” был введен английским математиком Дж.Дж. Сильвестром в 1878 году в статье, посвященной методам анализа структуры химических веществ, а первый учебник по теории графов был издан лишь в 1936 году. В течение двух столетий теория графов нашла применение в схемотехнике (схемы Г. Кирхгофа), инженерии, химии, социологии, урбанистике, геоинформационных системах и сети интернет.
Граф – это абстрактное представление множества объектов и связей между ними. Таким образом, математический граф описывается как совокупность ребер и вершин согласно формуле (1):
где G – граф; V – множество вершин; E – множество пар вершин, каждая из которых представляет собой описание связи (ребра) графа.Уровень абстракции, свойственный теории графов, достаточно высок: рассматриваются объекты (характеристики, параметры) лишь двух видов – вершины и соединяющие их ребра. Несмотря на это, данное представление оказывается полезным в столь различных отраслях, помимо названных, как информационный поиск (вершины – документы или веб-страницы, а ребра – ссылки между ними), лингвистика (взаимосвязь слов), теоретическая физика (диаграммы Фейнмана), логистика (расчет перевозок в транспортной сети), программирование (структуры данных), социальные науки (связи между отдельными людьми), молекулярная биология и экология, и многих иных. Впрочем, уровень абстракции может регулироваться: ребра могут рассматриваться как неориентированные, отражающие лишь факт связи, так и ориентированные, указывающие ее направленность; могут рассматриваться графы с несколькими ребрами (мультиграфы); ребра могут быть “нагруженными”, то есть им приписывается численное значение (например, пропускная способность, стоимость перевозок, мера статистической связи и другие). Особым направлением является изучение “случайных” графов, в которых ребра присутствуют с некоторой вероятностью. Значительный интерес могут представлять также численные характеристики структуры графа (Харари, 1973; Bornholdt, Schuster, 2003).
Основными преимуществами графового анализа являются:
1. Гибкость в выборе подходов к созданию графа (т.е. универсальность объекта исследования) и многообразие метрик, которые при этом можно использовать;
2. Привязка к топологии при решении пространственно зависимых задач;
3. Возможность использования различных подходов к атрибуции узлов и ребер;
4. Легкость представления графа в виде матрицы или, в случае некоторых специфичных графов, в виде кластера, что дает широкие возможности для применения самых разных математических подходов к обработке данных.
Следует отметить, что в 2021 году главная математическая премия – Абелевская – была присуждена за работы в области дискретной математики, в том числе – в области комбинаторики и графового анализа (Ustimenko, 2021).
Графовая теория активно развивается, разрабатываются новые подходы к решению различных задач (Kunszenti-Kovács et al., 2019; Viola, Wigderson, 2018). Такое внимание к теории графов и работам в этой области говорит о значимой роли данного раздела математики в компьютерных науках и обработке данных.
Общие сведения и показатели
Теория графов позволяет рассматривать головной мозг как сложную сеть или граф, состоящий из набора вершин (узлов) и связей между ними (ребер) (рис. 1). В качестве вершин могут выступать анатомические церебральные образования, а ребра соответствуют выявленным между ними связям. Тогда теоретико-графовый анализ будет иметь целью оценить структуру и степень связей различных областей мозга. Подобный подход дает возможность выделить основные компоненты сети и рассчитать ряд показателей, описывающих их свойства, а также выявить, какие вершины являются центральными (Ушаков, Величковский, 2015; Меринов, 2016; Faber et al., 2020).
Рис. 1.
Основные понятия теории графов (согласно О. Sporns, 2010 с дополнениями). Fig. 1. Basic concepts of graph theory (according to O. Sporns, 2010 with additions).
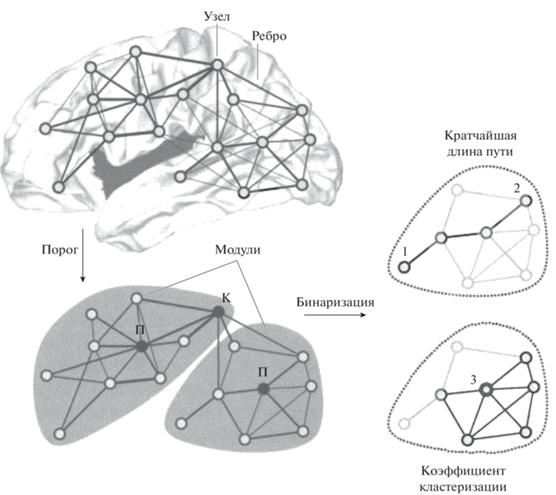
На рис. 1 демонстрируется ненаправленный взвешенный граф (с присвоенными ребрам условными значениями – весами) – до и после применения пороговой функции, которая убирает слабые связи (согласно Rubinov, Sporns, 2010; Меринов, 2016; Faber et al., 2020). Сеть состоит из двух модулей, соединенных хабом-коннектором (обозначен “К”), каждый из которых содержит по одному “провинциальному” хабу (обозначен “П”). Фрагменты справа иллюстрируют основные метрики графа после процедуры бинаризации (разбиения на отдельные модули): интеграции (кратчайшая длина пути между узлами 1 и 2) и сегрегации (локальный коэффициент кластеризации для узла 3. Узел 3 имеет 5 из 10 возможных связей между соседями и, cоответственно, коэффициент кластеризации 0.5).
Для изучения особенностей графа могут использоваться как физические метрики (например, длина тракта), так и топологические. Узлами графа могут являться регионы мозга, определенные на основе данных фМРТ, диффузионно-тензорной трактографии, а в качестве ребер – количественный или качественный показатель их взаимодействия (в направленных графах – оценка степени взаимовлияния нейросетей друг на друга, рассчитываемая через коэффициенты линейной и нелинейной каузальности по Грейнджеру, в простых – коэффициенты ковариации, когерентности и др.), т.е. анатомические или функциональные связи (Friston, 1994; Rubinov, Sporns, 2010; Dai et al., 2019), определяемые в публикациях последних лет как коннективность. Выделяют структурную, функциональную и эффективную коннективность (Friston, 1994; Meier et al., 2016; Sharaev et al., 2016; Sharova et al., 2018). Структурная коннективность описывает анатомические связи между участками головного мозга (проводящие пути). Функциональная коннективность отражает сходство характеристик паттерна нейрональной активности анатомически удаленных друг от друга образований головного мозга. Эффективная коннективность характеризует влияния одной зоны на другую (активация или торможение).
Этапы графового анализа
В практическом плане графовый анализ коннективности схематически можно разбить на несколько этапов. На первом выделяются основные составляющие части сети (вершины, узлы), то есть субъективные области интереса (region of interest, ROI). В зависимости от гипотезы для анализа могут быть выбраны одновременно все области мозга или отдельные регионы, разделенные в соответствии с международными стандартами (атлас Бродмана, AAL-атлас, MNI-пространство, атлас Талайраха (Dickey, 2017), система “10–20%” (“10–10%”) и другие (Koessler, 2009)). Далее измеряется активность внутри составляющих сети и связи между ними.
Наиболее универсальным вариантом расчета нагрузки ребер графа является вычисление коэффициентов корреляции Пирсона. Последний характеризует существование линейной зависимости между двумя величинами (Jang et al., 2018; Hasan et al., 2020). Для ГА ЭЭГ коэффициенты корреляции Пирсона рассчитываются между биопотенциалами заданных (или всех) отведений ЭЭГ. Наряду с этим для оценки нагрузки ребер возможно вычисление кросс-корреляции со сдвигом У относительно Х на время τ, что позволяет выявить сопряженность между сигналами, различающимися между электродами по времени появления (Русинов и др., 1987; Hassan et al., 2014). Другим распространенным вариантом является вычисление когерентности – количественного показателя, отражающего степень синхронизации определенных частотных составляющих спектра ЭЭГ между разными электродами (Русинов и др., 1987; Свидерская, 2008).
Следующий этап заключается в формальном анализе сетевой организации. На завершающем этапе параметры сети сравниваются для разных состояний и между разными группами испытуемых. Эти данные могут быть получены с помощью инвазивных или неинвазивных методов, как в парадигме, включающей наличие экспериментальной задачи, так и в условиях покоя.
Метрики для графового анализа
Исходя из представлений теории графов, в основе функциональной организации мозга лежат процессы сегрегации и интеграции (Friston, 2009), которые обеспечивают эффективную функциональную специализацию и глобальную коммуникацию. Оценить эти процессы можно, проанализировав глобальную и локальную коннективность мозга.
Функциональная сегрегация – процесс формирования локальных кластеров нейросети (наборов связанных между собой узлов). Основными метриками сегрегации являются коэффициент кластеризации и модульность.
Коэффициент кластеризации характеризует тенденцию к образованию групп взаимосвязанных узлов; для конкретного узла показывает, сколько ближайших соседей данного узла также являются ближайшими соседями друг для друга. Больший коэффициент кластеризации отражает большую сегрегацию (Rubinov, Sporns, 2010).
Модульность – это характеристика графа, связанная с возможностью выделить в нем отдельные кластеры (модули) (рис. 1), т.е. группы узлов, тесно связанных между собой при слабой связи между модулями (Newman, 2006; Bordier et al., 2017). Чем выше модульность, тем больше кластеров выделяется в функциональной сети мозга. Высокая кластеризация соответствует процессам локальной обработки информации.
Хабы – это функциональные узлы нейрональной сети, обладающие большим числом связей с соседями. Выделяют хабы-коннекторы (Sporns et al., 2007), соединяющие между собой разные модули графа (К на рис. 1), а также внутримодульные, или “провинциальные” хабы (П на рис. 1).
Функциональная интеграция – это способность быстро объединять специализированную информацию из распределенных областей мозга (Rubinov, Sporns, 2010). Оценить степень интеграции можно с помощью таких показателей, как характеристическая длина пути и глобальная эффективность.
Характеристическая длина пути – средняя длина кратчайшего пути между всеми парами вершин сети (Watts, Strogatz, 1998; Rubinov, Sporns, 2010). Кратчайшая длина пути – это минимальное количество ребер, соединяющих вершину со всеми другими вершинами. Она показывает, что от одного узла к другому можно дойти за малое число шагов (ребер).
Глобальная эффективность рассчитывается как среднее обратной матрицы расстояний. У полностью соединенной сети глобальная эффективность максимальна: все расстояния равны единице, все пары узлов связаны между собой. Короткая средняя длина кратчайшего пути и высокая глобальная эффективность означают более высокую интеграцию в мозге.
Таким образом, процесс сегрегации связан с функциональной специализацией областей мозга, в то время как процесс интеграции отвечает за объединение информации, полученной от разных регионов. Баланс между ними является важным условием оптимального функционирования распределенных нейронных сетей как основы реализации когнитивных функций и адаптивного поведения (Tononi et al., 1994; Fox, Friston, 2012).
Указанные процессы в значительной степени регулируются вниманием и сознанием. Сегрегация увеличивается в глубоком сне (Zhu et al., 2014), при простых сенсорных (зрительных) задачах (Maksimenko et al., 2018), в состоянии отдыха и при автоматизированных видах деятельности (Sporns, 2010). При выполнении сложных когнитивных заданий, включающих рабочую и эпизодическую память, когнитивный контроль, осознанное восприятие, напротив, возрастает интеграция, ключевая роль в которой принадлежит гиппокампу, префронтальной коре и гиппокампально-таламическим связям (Farahani et al., 2019).
Математический аппарат, используемый при работе с графами, не ограничивается указанными выше характеристиками. К числу перспективных относят, в частности, анализ графов посредством нахождения “минимального остовного дерево” (spanning tree) – подмножества ребер графа, которое соединяет все вершины с минимальным весом (т.е. минимальной суммой весов ребер). Это важно, например, для нахождения кратчайшего пути с единственным источником (Ion et al., 2006). В табл. 1 собраны основные метрики ГА, которые сегодня обсуждаются в научной литературе (Vecchio et al., 2017, Sporns, 2018; Dell’Italia, 2018; Фекличева и др., 2020; Faber et al., 2020; Liégeois et al., 2020).
Таблица 1.
Графовые метрики Table 1. Graph metrics
| Метрики | Основные определения | Ссылки |
|---|---|---|
| Базовые метрики (Basic metrics) | ||
| Степень узла (Node degree) |
Число связей узла с другими узлами, равно количеству соседей узла. Степени всех узлов – маркер развития и устойчивости сети. Средняя степень сети – метрика плотности или общих “затрат на проведение” |
(Rubinov, Sporns, 2010) |
| Кратчайшая длина пути (Shortest path length) |
Кратчайшее расстояние между узлами i и j | (Rubinov, Sporns, 2010) |
| Число треугольников (Number of triangles) |
Число треугольников вокруг узла i | (Rubinov, Sporns,2010) |
| Метрики интеграции (Integration metrics) | ||
| Характеристическая длина пути (Characteristic path length) |
Средняя кратчайшая длина пути для всех пар узлов в сети | (Watts, Strogatz, 1998) |
| Глобальная эффективность (Global efficiency) |
Величина, обратная характеристической длине пути | (Latora, Marchiori, 2001) |
| Метрики сегрегации (Segregation metrics) | ||
| Локальный коэффициент
кластеризации (Local clustering coefficient) |
Отношение числа связей между соседями узла к возможному числу связей между соседями; характеризует, насколько хорошо связаны между собой соседи данного узла | (Watts, Strogatz, 1998) (Onnela, 2005) (Fagiolo, 2007) (Saramaki et al., 2007) |
| Глобальный коэффициент
кластеризации (Global clustering coefficient) |
Значения кластеризации для всех узлов графа. Высокое значение этого показателя свидетельствует о том, что узлы плотно сгруппированы в пределах графа; низкое – что связи в графе относительно равномерно распределены среди всех узлов | |
| Транзитивность (Transitivity) |
Отношение между наблюдаемым числом закрытых триплетов и их максимально возможным числом в графе. Выявляет наличие плотно соединенных сообществ (кластеров). Сети малого мира, как правило, характеризуются высокой транзитивностью | (Newman, 2003) |
| Локальная эффективность узла (Local efficiency) |
Величина, обратная среднему кратчайшему пути для всех соседей данного узла | (Latora, Marchiori, 2001) |
| Модульность (Modularity) |
Метрика, показывающая подразделение сети на модули. Сети с высокой модульностью обладают плотными связями между узлами внутри модулей, но разреженными – между узлами в различных модулях. Выявление модульности часто используется как способ оптимизации для определения структуры сообщества | (Newman, 2006) |
| Метрики центральности (Centrality metrics) | ||
| Степень близости узла (Closeness centrality) |
Мера центральности, вычисляемая как обратная величина суммы длин кратчайших путей между узлом и всеми другими узлами графа; чем более централен узел, тем ближе он ко всем другим узлам | (Freeman, 1978), (Rubinov, Sporns, 2010) |
| Степень посредничества (Betweenness centrality) |
Мера центральности в графе, основанная на кратчайших путях. Для любой пары вершин в связном графе существует по меньшей мере один кратчайший путь между вершинами, для которого минимально либо число ребер, по которым путь проходит (для невзвешенных графов), либо сумма весов этих ребер (для взвешенных графов). Степень посредничества для каждой вершины равна числу кратчайших путей через вершину. Узел с наивысшей степенью посредничества имеет больший контроль сети, поскольку больше информации проходит через него | |
В случае связанности между сигналами, полученными со всех точек измерения (будь то области интереса фМРТ, характеристики трактографии либо отведения ЭЭГ), получается так называемый полный граф (все вершины соединены друг с другом). Именно полная связность является формальным требованием при определении нескольких глобальных метрик графа (Hatlestad-Hall et al., 2021). Вместе с тем для расчета ряда топологических характеристик, приведенных в табл. 1, слабые связи должны быть отброшены. Для того чтобы удалить слабые или статистически незначимые взаимодействия, используются различные методы статистического анализа (Храмов и др., 2021). Следует отметить, что этот весьма важный вопрос подбора порога более детально проанализирован в модельных исследованиях (Bordier et al., 2017) и в работах по нейровизуализации (фМРТ, трактография) (Buchanan et al., 2020; McColgan et al., 2018). Так, согласно (McColgan et al., 2018), можно применять как минимум два метода его оценки: абсолютный и относительный порог. В качестве абсолютного порога используются нормализованные значения силы связи; чем выше абсолютный порог, тем разреженнее граф. Для определения относительного порога оцениваются соотношения сильных связей в графе; чем ниже относительный порог, тем разреженнее граф. Специальные опции для выбора абсолютного или относительного порога содержатся в ряде универсальных пакетов программ для графового анализа, как например GraphVAR (см. табл. 2).
Таблица 2.
Программы для графового анализа данных ЭЭГ и нейровизуализации Table 2. Programs for graph analysis of EEG data and neurovisualization
| Программы | Данные для анализа | Область применения |
|---|---|---|
| Python | ||
| Python MNE | МЭГ, ЭЭГ, стереоЭЭГ, ЭКоГ, БИКС | Графовый анализ данных и нейровизуализация, с локализацией источников |
| Python NetworkX | МЭГ, ЭЭГ, стереоЭЭГ, ЭКоГ, БИКС | Графовый анализ данных и нейровизуализация; для локализации источников необходимы дополнительные программы (напр., Python MNE) |
| Matlab | ||
| Brain Connectivity Toolbox | МРТ (диффузионная, структурная, функциональная), МЭГ, ЭЭГ | Графовый анализ данных, для локализации источников необходимы дополнительные программы (напр., ЕЕGLAB) |
| GraphVAR (комплексный тулбокс, включающий в себя: Brain Connectivity Toolbox, Network Based Statistic, BRAPH, lassBrainNetC) | МРТ (диффузионная, структурная, функциональная), МЭГ, ЭЭГ | Графовый анализ данных, для локализации источников необходимы дополнительные программы (напр., EEGLAB) |
| BrainStorm | МРТ, МЭГ, ЭЭГ, БИКС, ЭКоГ, глубинные электроды | Анализ нейрофизиологических данных и нейровизуализация |
| BrainNetViewer | – | Нейровизуализация |
| Network-Based Statistic (NBS) | МРТ (диффузионная, структурная, функциональная), ЭЭГ (данные должны быть конвертированы) | Статистический анализ |
| Statistical Parametric Mapping (SPM) | фМРТ, ПЭТ, ЭЭГ, МЭГ | Статистический анализ |
| Отдельная программа (распространяется свободно) | ||
| sLORETA | ЭЭГ (данные должны быть конвертированы в ascii формат) | Локализация распределенных источников |
Визуализация результатов графового анализа
На рис. 2 представлены варианты отображения результатов ГА, которые можно использовать применительно к данным, полученным разными методами исследования (фМРТ, ЭЭГ и др.). Наиболее простая и универсальная форма – это двумерный график, на котором по оси У – значения одной из глобальных характеристик графового анализа – интеграции или сегрегации (средняя кратчайшая длина пути, степень посредничества и т.д.), а по оси Х – параметры: например, частотный диапазон ЭЭГ (рис. 2 (а)). На рис. 2 (а) показан пример трактовки результатов расчета среднего коэффициента кластеризации. Для двух условий эксперимента (например, состояния покоя/выполнения движения, состояния покоя с открытыми/закрытыми глазами и др.) по каждому ритму ЭЭГ рассчитывается средний коэффициент кластеризации сети (среднее локальных коэффициентов кластеризации для всех узлов). Значения этого коэффициента при исследуемых состояниях сравниваются между собой. Диаграмма на рисунке дает представление о том, как изменяется связность узлов по каждому ритму в зависимости от проводимого эксперимента. Высокий средний коэффициент кластеризации сети свидетельствует о большом числе взаимосвязей между узлами в сети, тенденции к образованию групп узлов. Однако в случае подобного представления данных возможности анализа существенно обеднены отсутствием информации о топографических особенностях коннективности исследуемых состояний – наряду с глобальными метриками сегреграции и интеграции существует большой набор паттернов локальной коннективности. Метрики сегрегации могут быть основаны на подсчете треугольников, но часто включают также более сложную декомпозицию на модули (Szalkai et al., 2017).
Рис. 2.
Варианты отображения результатов графового анализа. (а) – на основе базовых метрик: изменения среднего коэффициента кластеризации (сКК) по диапазонам физиологических ритмов ЭЭГ в зависимости от двух вариантов эксперимента: синяя линия – вариант 1, красная – вариант 2. (б) – по сочетанию глобальных и локальных метрик: слева – модули (голубой фон), кратчайший путь между узлами А и Б (охровый), треугольник (зеленый); справа – степень узла (фиолетовый), хаб (желтый), мотивы (оранжевый, выделены сплошной (функциональный мотив) и пунктирной (анатомический мотив) линиями). Fig. 2. Options for displaying the results of graph analysis. (а) – basic metrics: average clustering coefficient in different frequency ranges for two experiment conditions: blue line – option 1, red – option 2. (б) – global and local metrics: left – modules (blue), shortest path length between nodes A and Б (ocher), triangle (green); right – degree (purple), hub (yellow), motifs (orange; indicated by solid line (functional motif) and dotted line (anatomical motif).
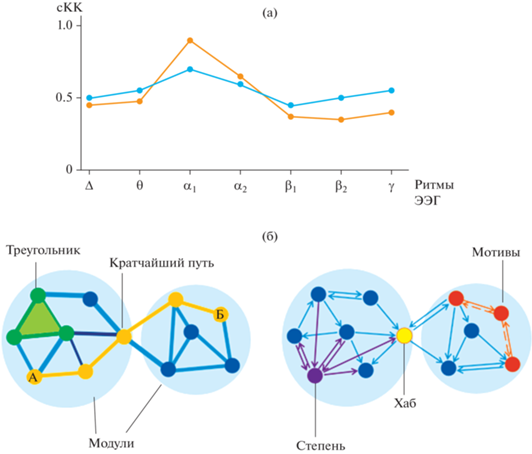
Сочетание представленности глобальных и локальных характеристик ГА показано на рис. 2 (б). Зачастую образующиеся взаимосвязи между узлами называют анатомическими треугольниками, если речь идет о трех узлах. Если в таких треугольниках присутствуют бинаправленные связи между узлами, то их называют “мотивами” (см. рис. 2 (б)). На сегодня определение направленности связей является отдельной задачей при проведении графового анализа данных фМРТ или ЭЭГ (Sharaev et al., 2016), для этого используют уже априори известные анатомические или причинные связи. Направленность связей выявляют также посредством анализа изменений функциональной активности, которые возникают вследствие локальных пертурбаций (Rubinov, Sporns, 2010).
Если какой-либо из мотивов проявляется при анализе экспериментальных данных с большей частотой, чем остальные, то говорят, что он более значим. Как правило, при определении значимости используют условие случайной нулевой гипотезы, то есть выбирается некая случайная сеть, в которой априори не может возникать мотивов. При этом для определенного узла можно рассматривать как частоту возникновения определенного мотива, так и в целом частоту возникновения любых мотивов, связанных с этим узлом. Высокая частота возникновения мотивов говорит о значимости узла при выполнении проводимого эксперимента (Onnela et al., 2005).
Рис. 3.
Вариабельность топологической организации сетей. 1 – регулярный граф, 2 – граф “малого мира”, 3 – случайный граф. Линия со стрелкой – рост случайности графа. Fig. 3. Variability in the topological organization of networks. 1 – regular graph, 2 – graphs with small-world properties, 3 – random graph. The line with an arrow is the increase in the randomness of the graph.
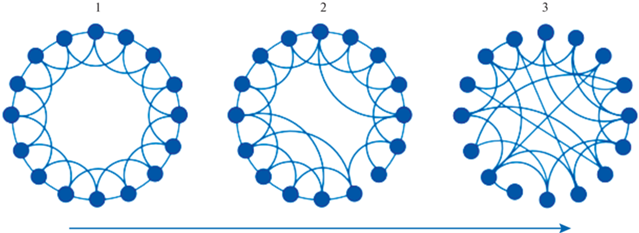
Вариабельность топологической организации сетей
Топология сетей может варьировать от полностью случайной до полностью упорядоченной (решетка) (риc. 3). В работе (Watts, Strogatz, 1998) исследованы некоторые динамические системы (такие как нервная система нематоды, электрическая сеть, сеть на основе данных об актерах кино) и показано, что, помимо обычного разделения на регулярные и случайные сети, можно дополнительно выделить сети малого мира. Они кластеризованы в большей степени, чем случайные сети, но имеют примерно такую же длину пути, как и случайные (Watts, Strogatz, 1998; Hagmann et al., 2007; Меринов, 2016; Faber et al., 2020). Сети малого мира имеют модульную структуру с плотным внутримодульным взаимодействием и оптимальное число межмодульных соединений (Gong et al., 2009; Hagmann et al., 2007).
Было обнаружено, что анатомические сети мозга кошки и макаки (Hilgetag, Kaiser 2004; Sporns, Zwi, 2004; Hilgetag et al., 2019), а также анатомические и функциональные сети мозга человека (Bassett et al., 2006; Vaessen et al., 2010) соответствуют структуре малого мира. Такая структура обеспечивает высокую эффективность локальной и глобальной передачи информации (из одной вершины сети в любую другую вершину, не смежную с ней, можно попасть за небольшое количество шагов), а также способствует параллельной обработке в иерархически организованных модулях (Bullmore, Sporns, 2009; Bassett, Bullmore, 2017). При этом важно подчеркнуть, что для коннектомов макак и человека решающее значение имеет эволюция с сохранением локальной кластеризации, в то время как для примитивных организмов достаточно сохранения усредненной кластеризации (Pospelov et al., 2019).
Структура малого мира была признана идеальной организационной архитектурой, что выражается в небольших затратах энергии на передачу информации и оптимальном балансе между глобальной интеграцией и локальной обработкой информации (Tan et al., 2013; Liao et al., 2017). С помощью моделей было рассчитано, что мозг организован по принципу “мир тесен”: характеризуется высоким коэффициентом кластеризации и малой средней кратчайшей длиной пути. Это соответствует варианту организации “экономичной” сети малого мира, связанной с общепринятыми понятиями цены и качества (Latora, Marchiori 2001; Bullmore, Sporns, 2012). Топологически кластеризованные узлы и анатомически, предположительно, должны быть расположены рядом, так как это позволяет минимизировать энергетические затраты на передачу сигналов. Подобная сеть обеспечивает более высокую скорость обработки информации и обучения (Simard et al., 2005). Сети малого мира характеризуются высоким уровнем локальной кластеризации и ограниченным числом случайных соединений на большие расстояния (Watts, Strogatz, 1998; Latora, Marchiori, 2001; Sporns, 2004, 2018). Эта организация нарушается при церебральной патологии и имеет тенденцию к восстановлению при позитивной клинической динамике.
Индекс малого мира (S) – показатель, позволяющий количественно оценить степень соответствия сетевой организации с прототипом архитектуры малого мира. Математически он представляет собой отношение между локальной связностью и глобальной интеграцией сети. Показатель малого мира обычно рассматривается как мера оптимальной эффективности конфигурации сети.
Математически S определяется как:
где L – длина пути заданного графа;
C – коэффициент кластеризации заданного графа;
L0 – длина пути cлучайного графа;
C0 – коэффициент кластеризации случайного графа.
Сеть обладает свойствами “малого мира”, когда S > 1 (Humphries, Gurney, 2008). Чтобы оценить граф на соответствие структуре малого мира, его обычно сравнивают со структурой случайно сгенерированного графа. Случайный граф – это граф, свойства которого определяются некоторым случайным образом. Граф обладает структурой “малого мира”, если его длина пути подобна длине случайного графа: L ≥ L0, тогда как коэффициент кластеризации намного больше, чем у случайного графа: C $ \gg $ C0.
Графовый анализ в нейровизуализации
В нейронауках графовый подход первоначально был применен к анализу данных нейровизуализации (фМРТ, ПЭТ, трактография) (Friston, 1994; Hagmann et al., 2007; Gong et al., 2009; van den Heuvel, Hulshoff Pol, 2010; Wu et al., 2018). Затем метод стал успешно использоваться для анализа многоканальных записей ЭЭГ и МЭГ здоровых людей и пациентов с разными формами церебральной дисфункции (Bassett et al., 2006; Chennu et al., 2014).
Привлечение ГА способствовало выявлению различных свойств и топологических характеристик фМРТ-сетей мозга. При этом глобальная эффективность, минимальная длина пути, степень посредничества использованы как метрики функциональной интеграции, для оценки распространения информации в сети на глобальном уровне (Termenon et al., 2016), а локальная эффективность и коэффициент кластеризации – как метрики функциональной сегрегации, для оценки передачи информации на локальном уровне (Rubinov, Sporns, 2010). Установлено, что процессы сенсомоторной и когнитивной обработки в мозге могут быть как локализованы в специализированных регионах, так и распределены в крупномасштабных сетях и представлены когерентными осцилляциями. Таким образом, функциональная архитектура мозга может рассматриваться в тесной связи с этими процессами (Bassett, Sporns, 2017; Shine et al., 2019). Причем архитектура глобальных нейронных сетей в существенной степени является наследуемой (Лисичев и др., 2020)
По данным литературы, анатомическая коннективность в первых исследованиях на здоровых испытуемых изучалась с применением косвенных методов – расчета статистически значимых корреляций между показателями толщины коры, полученными с помощью структурной МРТ (He et al., 2007; Chen et al., 2008). Было показано, что анатомическая организация мозга обладает свойствами сети малого мира (низкая средняя кратчайшая длина пути, высокий глобальный коэффициент кластеризации). Кроме того, для нее характерно присутствие модулей, которые соответствовали хорошо известным из литературы функциональным сетям: языковой, исполнительной, сети памяти, визуальной и сенсомоторной (Chen et al., 2008). Сходные результаты были получены с применением диффузионно-тензорной визуализации и диффузионно-спектральной томографии (Hagmann et al., 2008, 2007; Iturria-Medina et al., 2007, 2008; Gong et al., 2009).
В ряде исследований было обнаружено, что функцию хабов-коннекторов (участков, обладающих наибольшим количеством анатомических связей с другими областями) выполняют префронтальные, париетальные и височные отделы полушарий. Рассматривая совокупность результатов, можно отметить, что выявленные хабы-коннекторы частично перекрываются (особенно в задних корковых областях) с дефолтной системой мозга и с сетями, ответственными за процессы внимания (Guye et al., 2010; Wang et al., 2010). При этом, согласно литературе, хабы-коннекторы характеризуются высоким уровнем экспрессии генов, важных для когнитивных функций (Rubinov et al., 2015) и процессов межнейронной коммуникации, таких как глутамат- и ГАМК-эргическая передача (Arnatkeviciūtė et al., 2018).
В работе с применением комбинации методов диффузионно-спектральной томографии и фМРТ вышеупомянутые задние корковые регионы, в особенности предклинье и задние поясные, обозначены как “структурное ядро” коры головного мозга (Hagmann et al., 2008). Данные области демонстрируют высокий уровень метаболической активности как в состоянии покоя, так и при предъявлении специальных заданий. Считается, что именно здесь пересекаются процессы обработки информации разных модальностей. Известно, что указанные зоны мозга поражаются при нейродегенеративных заболеваниях, например, болезни Альцгеймера (Buckner et al., 2009). Повреждение этих участков при патологических состояниях может отразиться на состоянии высших психических функций. В этой связи представляется важным исследовать как анатомические, так и функциональные особенности тех церебральных областей, которые характеризуются максимальным числом связей и оказывают наиболее существенное влияние на общую сетевую организацию.
Было выявлено, что в процессе онтогенеза топологическая организация сетей головного мозга человека изменяется по обратной U-образной траектории: на ранних стадиях развития наблюдается рост интегрированной топологии с выходом на относительное плато во взрослой жизни, но ростом более локализованной топологии в поздний период жизни (Collin, van den Heuvel, 2013). Уточняя эти данные, исследование (Stumme et al., 2020), подтверждающее общую тенденцию к ослаблению межсетевой и внутрисетевой интеграции (прежде всего для областей первичной обработки информации) в пожилом возрасте, демонстрирует возможное усиление на этом фоне взаимодействий между сетями более высокого порядка, особенно с сенсомоторной. Эти системные перестройки интерпретируются авторами как компенсаторная попытка поддержания когнитивных функций за счет интеграции механизмов контроля. В целом, результаты ГА позволяют полагать, что в эволюционном плане архитектура малого мира была развита, чтобы повысить сложность или адаптивность функций головного мозга, а также минимизировать затраты энергии (Bassett, Sporns, 2017).
Нейросетевая организация нарушается при аномалиях развития (Chen et al., 2019), при эпилепсии (Bernhardt et al., 2015), а также при различных pасстройствах психики (Fornito, Bullmore, 2015; Finotelli et al., 2018). Снижение глобальной и локальной эффективности выявлено ГА фМРТ при рассеянном склерозе, что позволяет классифицировать это заболевание как синдром дисконнективности (Farahani et al., 2019).
Согласно результатам ГА фМРТ как в состоянии покоя, так и при выполнении заданий, ухудшение некоторых когнитивных функций, связанное с физиологическим (Wang et al., 2010) и патологическим (болезнь Альцгеймера) (Phillips et al., 2015; Dai et al., 2019) старением, сопряжено со снижением индекса малого мира и повышенной случайностью функциональных сетей. При этом у пациентов с болезнью Альцгеймера локализация главных нейросетевых хабов, включающих заднюю поясную кору, предклинье, нижнюю теменную долю и медиальную фронтальную кору и предположительно задействованных в прогрессировании болезни, коррелирует с расположением церебральных зон отложения бета-амилоида (Farahani et al., 2019).
Напротив, при обратном развитии патологического процесса, как, например, повышение уровня сознания, угнетенного после тяжелой черепно-мозговой травмы, по данным ГА фМРТ покоя отмечается тенденция к восстановлению нейросетевой организации с приближением по структуре к сети малого мира (Dell’Italia et al., 2018).
Приведенные данные свидетельствуют о том, что более детальное изучение особенностей нейросетевых изменений при разных формах церебральной патологии позволяет расширить понимание биологических механизмов развития заболеваний (Douw et al., 2019; Demertzi et al., 2019).
Варианты графового анализа ЭЭГ
Графовый анализ ЭЭГ получает все большее распространение при исследовании особенностей сетевой организации здорового и больного мозга в последние годы. Подготовка к ГА данных ЭЭГ заключается в представлении результатов оценки биоэлектрических сигналов в виде матриц смежности, где узлами являются позиции электродов, ребрами – показатели функциональной коннективности (см. раздел “Этапы графового анализа”). В этом качестве могут привлекаться показатели корреляции, когерентности, вейвлет-синхронности и иные, используемые в электроэнцефалографии. Вопрос о выборе порога коннективности для ГА ЭЭГ освещен в литературе менее детально (Hatlestad-Hall et al., 2021) по сравнению с публикациями по нейровизуализации (см. раздел “Метрики для графового анализа”). Полагаем, что при его решении посредством статистических методов представляется целесообразным учитывать также опубликованные данные о наиболее характерных для здоровых людей показателях коннективности ЭЭГ (как например, Русинов и др., 1987), либо результаты собственных исследований в контрольных группах.
Методические подходы, используемые для графового анализа ЭЭГ, достаточно вариативны. Более традиционным и относительно простым представляется ГА конвекситальных биопотенциалов, основанный, как и в случае гемодинамических сигналов (Liégeois et al., 2014), на вычислении матрицы их корреляций – без первичного нахождения источников генерации составляющих ЭЭГ. Связанный с таким подходом вопрос о механизмах распространения электрического поля в мозге как в объемном проводнике достаточно исследован теоретически и экспериментально (Егоров, Кузнецова, 1976; Nunez, Srinivazan, 2006). К настоящему времени разработаны способы подавления эффекта объемной проводимости (Храмов и др., 2021). Кроме того, получаемые с помощью данного подхода результаты подтверждаются опытом электроэнцефалографии, прежде всего методами локализации источников (Гнездицкий, 2004; Pascual-Marqui et al., 2021).
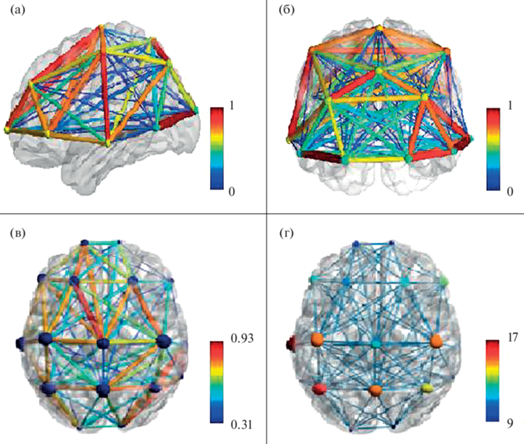
К числу распространенных программных пакетов (toolbox), используемых для указанной модификации ГА ЭЭГ, относится BrainNet Viewer (Xia et al., 2013). Причем возможно проведение расчета графовых показателей по формулам на базе одного программного пакета, а выведение их в виде графиков – на базе другого. Так, на рис. 4 представлен пример визуализации ГА-показателей корреляции биопотенциалов при монополярной 18-канальной записи ЭЭГ здорового испытуемого в состоянии спокойного бодрствования с закрытыми глазами. Первоначально в среде MatLab рассчитывалась матрица коэффициентов корреляции Пирсона (ребра графа). Полученные результаты визуализировались в программном обеспечении BrainNetViewer. При этом возможно отображение как силы взаимодействия (рис. 4 (в)), так и активности отдельных узлов графа (рис. 4 (г)). На рис. 4 (a) и 4 (б) приведены примеры возможного отображения результатов в различных проекциях.
Рис. 4.
Примеры визуализации показателей корреляции ЭЭГ в программном пакете BrainNetViewer (данные В.А. Бордюг). (a) и (б) – схема возможного отображения поверхности, ребер и узлов графа: в сагиттальной (a) и фронтальной (б) проекциях. (в) и (г) – данные ГА ЭЭГ здорового испытуемого 23 лет в состоянии спокойного бодрствования с закрытыми глазами в аксиальной проекции; (в) – ребра графа – уровень корреляции биопотенциалов между отведениями ЭЭГ, обозначенный цветовой шкалой справа; узлы графа – отведения ЭЭГ; (г) – ребра графа – уровень корреляции биопотенциалов между отведениями; узлы графа – степень узла (число связей с другими узлами), обозначенная цветовой шкалой справа. Fig. 4. Examples of visialization of EEG correlation coefficients in BrainNetViewer (data of V.A. Bordiug). (a) and (б) – scheme of possible visualization of graph surface, nodes, and edges: in saggital (a) and frontal (б) projections. (в) and (г) – data of GA EEG for healthy subject (man, age – 23), quiet wakefulness with eyes closed in axial projection; (в) graph edges – correlation coefficient between EEG channels, indicated by colorscale on the right; graph nodes – EEG channels; (г) graph edges – correlation coefficient between EEG channels; graph nodes – node degree (number of its links), indicated by colorscale on the right.
Посредством такого подхода к оценке ЭЭГ была показана онтогенетическая динамика организации сетей малого мира мозга человека (Tóth et al., 2017), включая переход от случайных к более организованным функциональным сетям малого мира при нормальном созревании мозга (Boersma et al., 2011).
ГА корковых биопотенциалов способствует исследованию взаимосвязи когнитивной активности и функциональной коннективности, сочетая анализ показателей глобальной нейрональной динамики и частотных характеристик отдельных церебральных регионов. Так, у здоровых взрослых испытуемых параметры кластеризации дельта-, тета- и гамма-диапазонов ГА МЭГ в состоянии покоя коррелируют с когнитивными показателями их тестирования. Отмеченная при этом меньшая кластеризация и более короткая длина пути в дельта-диапазоне у женщин указывает, по мнению авторов, на большую эффективность рабочей нейросетевой архитектуры женского мозга (Douw et al., 2011). В обзоре (Фекличевa и др., 2020) приводятся данные о наличии положительной корреляции коэффициента кластеризации и числа ребер графа ЭЭГ покоя, но значимо отрицательной – для длины пути с уровнем невербального интеллекта.
При выполнении задания с вовлечением рабочей памяти ГА ЭЭГ здоровых лиц выявил соответствие структуре малого мира показателей в альфа-, бета- и гамма-диапазонах (Micheloyannis et al., 2006). Кроме того, во время решения подобных задач значимо увеличивалась глобальная эффективность в тета-диапазоне и снижался коэффициент кластеризации в альфа. Отмечены определенные пространственные изменения коэффициента центральности тета-ритма: его снижение в левых средней лобной, нижней затылочной и язычной извилинах при увеличении в правой и левой прецентральных извилинах (Dai et al., 2017).
Определены нарушения (отличия от нормы) графовых показателей ЭЭГ, характерные для некоторых форм церебральной патологии: мигрень (Cao et al., 2016), шизофрения (Micheloyannis et al., 2006; Olejarczyk, Jernajczyk, 2017), аутизм (Zeng et al., 2017). Например, у пациентов с шизофренией наибольшие изменения параметров ГА по сравнению со здоровыми людьми выявлены для альфа-диапазона ЭЭГ: снижение силы связанности, уменьшение коэффициентов кластеризации и длины пути. Это сопровождается редукцией количества потоков в лобных корковых областях наряду с их увеличением от лобных отделов полушарий к затылочным (Olejarczyk, Jernajczyk, 2017). Последнее согласуется с гипотезой (Micheloyannis, 2018) о гипофронтальности при шизофрении, которая может быть обусловлена в том числе и нарушениями лобно-таламического взаимодействия (Finotelli et al., 2019).
ГА ЭЭГ детей с аутизмом (в возрасте 8–13 лет), проведенный (Zeng et al., 2017), показал значительное ослабление глобальных церебральных связей тета- и альфа-активности по сравнению с нормально развитыми детьми. У аутистов обнаружено уменьшение длины пути вместе с выраженным снижением коэффициента кластеризации в альфа-диапазоне, что указывало на нарушение архитектуры малого мира (снижение его индекса) и повышение случайности функциональных сетей. По мнению авторов, обнаруженные нейросетевые нарушения могут служить биомаркерами при диагностике данной патологии.
Выявлен ряд особенностей пространственной организации ритмов ЭЭГ при разном уровне патологического угнетения сознания (Chennu et al., 2014). Отмечена информативность оценки нейросетей ЭЭГ при прогнозировании его восстановления (Chennu et al., 2017; Carrasco-Gómez et al., 2021). Так, в работе (Chennu et al., 2017) показано, что сетевая метрика, индексирующая наличие плотно связанных между собой центральных узлов с высоким коэффициентом участия лобных и теменных корковых областей, касающаяся прежде всего характеристик ГА альфа-диапазона, распознавала текущий уровень сознания, угнетенного после черепно-мозговой травмы, и перспективы его восстановления с точностью, сравнимой с экспертной оценкой на основе позитронно-эмиссионной томографии.
В ряде публикаций по ГА ЭЭГ эффективно используется показатель “минимальное остовное дерево”. Его применение в онтогенетических исследованиях позволило выявить выраженные изменения функциональной коннективности мозга ребенка в возрасте около 7 лет (Boersma et al., 2013). ГА отдельных ритмов ЭЭГ способствовал выявлению возможных индивидуальных специфических реакций мозга на различные варианты лечения (Crobe et al., 2016). В качестве перспективного направления применения “минимальных остовных деревьев” является определение эмоциональных состояний. В недавних работах (Cao et al., 2020; Farashi, 2020) точность их верификации по данным ЭЭГ превышала 80%.
Другим активно развиваемым методическим направлением является графовый анализ сигналов ЭЭГ, основанный на предварительной локализации источников исследуемых биопотенциалов. Было показано, что функциональные сети, вычисляемые по данным конвекситальной ЭЭГ и от рассчитанных источников ее генерации, все же не идентичны (Perazaa et al., 2012). Это может быть обусловлено искажением конвекситальных сигналов нейронного происхождения свойствами проводящих сред, а также их смешением с немозговыми физиологическими сигналами (КГР, ЭКГ, дыхательные волны и др.).
Расчет локализации нейронных источников электрической активности по измерению потенциалов со скальпа называется “обратной задачей ЭЭГ” (Гутман, 1980; Nunez, 1981; Гнездицкий, 2004; Awan et al., 2019). Наиболее корректно решение обратной задачи сводится к тому, чтобы по распределению конвекситальных электрических потенциалов, измеренных одновременно в нескольких точках с известными координатами, установить источники генерации, их мощность, ориентацию и месторасположение в объеме головного мозга. Одной из модификаций данного подхода является анализ распределенных источников, согласно которому признается, что все источники биопотенциалов одновременно активны в той или иной степени. Наиболее популярными моделями распределенных источников, используемыми в настоящее время при анализе ЭЭГ, являются модификации метода взвешенной минимальной нормы (WMN) (Jatoi, Kamel, 2017; Michel, Brunet, 2019).
Данный подход активно разрабатывается и используется в исследованиях, руководимых P.M. Rossini (Vecchio, Miraglia, Rossini, 2017; Vecchio et al., 2017; Hatlestad-Hall et al., 2021). Наиболее поздняя разновидность реализованной на его основе программы – sLORETA (стандартизованная электромагнитная томография мозга низкого разрешения) основана на допущении, что регистрируемая на поверхности головы активность генерируется большими популяциями близко расположенных нейронов (Pascual-Marqui et al., 2002, 2011, 2021). Программа находится в свободном доступе и не требует установки MatLab. Алгоритм позволяет по распределению потенциалов на поверхности головы математически рассчитать локализацию источников электрической активности в объеме головного мозга с последующей визуализацией. Для этого в программе используется трехслойная сферическая модель головы, соотнесенная с оцифрованным атласом Talairah and Tournoux (1998, Brain Imaging centre, Montreal Neurological Institute). В sLORETA пространство ограничено серым веществом и областью гиппокампа, где предположительно сгруппированы основные активные источники.
Активность источников усредняется в пределах выбранных исследователем областей интереса (ROI, regions of interest) или в пределах областей Бродмана. Далее рассчитываются функциональные связи между этими областями (в виде матрицы значений размером N на N, где N – область мозга). Полученные значения коннективности можно использовать для последующей визуализации сети как графа с использованием дополнительных приложений или скриптов.
К числу вариантов представления результатов пространственного анализа ЭЭГ в программе sLORETA относится отображение локализации внутримозговых источников биоэлектрической активности (рис. 5 (а)), а также топографических особенностей функциональной коннективности отдельных корковых зон (выступающих в качестве вершин графа) по числу устанавливаемых связей (рис. 5 (б)).
Рис. 5.
Примеры визуализации результатов графового анализа ЭЭГ с предварительной локализацией источников активности. Результаты двухфакторного дисперсионного анализа (данные Е.А. Прошиной). (a) – плотность тока источников бета3 активности. (б) – связи ЭЭГ в дельта и альфа-2 диапазонах, преобладающие у здоровых испытуемых с генотипом LL относительно SL + SS полиморфизмом гена SLC6A4 (красные окружности символизируют центроид ROI (т.е. области Бродмана), размер окружностей пропорционален значению степени, т.е. количеству связей этой области; изображение получено на основе визуализации данных sLORETA в приложении BrainNetViewer). Fig. 5. Visualization examples of EEG data graph analysis with preliminary source localization. Results of two-way ANOVA test (data of E.A. Proshina). (a) – current density of sources of beta3 activity. (б) – EEG connections in the delta and alpha-2 ranges, which predominate in healthy subjects with the LL genotype relative to the SL + SS polymorphism of the SLC6A4 gene (red circles symbolize the ROI centroid (i.e. Brodmann’s area), the size of the circles is proportional to the degree value, i.e. the number of connections in this area; image derived from sLORETA data visualization in BrainNetViewer).
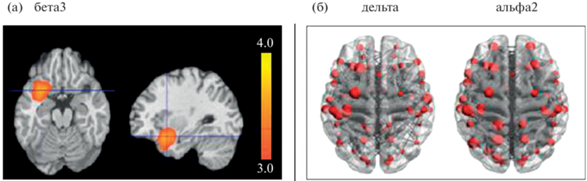
Описываемый подход был успешно использован для поиска ЭЭГ-маркеров болезни Альцгеймера (Vecchio, Miraglia, Rossini, 2017; Franciotti et al., 2019; Babiloni et al., 2020), для выявления различий между физиологическими и патологическими признаками старения головного мозга (Knyazev et al., 2015; Miraglia et al., 2016), для обнаружения глобальных и локальных нейросетевых нарушений, характерных для пациентов с фокальной эпилепсией (Hatlestad-Hall et al., 2021), определения нарушений коннективности ЭЭГ, специфичных для разных форм угнетения сознания при церебральной патологии (Cacciola et al., 2019).
На базе программы sLORETA доступны разные алгоритмы вычисления коннективности – с их возможной дальнейшей модификацией. В частности, Bosch-Bayard с соавт. был предложен расчет так называемой эффективной (направленной) когерентности (iCoh) с последующим результативным привлечением этого показателя для анализа особенностей сетевой организации ЭЭГ в норме (при закрытых и открытых глазах) и при церебральной дисфункции (дислексия) (Pascual-Marqui et al., 2014; Bosch-Bayard et al., 2020). Данные из sLORETA могут быть экспортированы и использованы в работе с другими специализированными приложениями, включая различные методы моделирования.
Следует отметить, что возможность предварительной локализации источников исследуемых биопотенциалов с последующей оценкой функциональных связей между ними или иными заданными областями интереса реализованы и в других современных программных приложениях: BESA, Brainstorm, OpenMEEG и т.д. Полученные показатели коннективности могут быть использованы для дальнейшего графового анализа. В этом контексте приведенные выше примеры ГА ЭЭГ на базе sLORETA лишь иллюстрируют возможности подобного подхода.
В табл. 2 приводятся наиболее распространенные пакеты программ, используемые для графового анализа данных ЭЭГ (с локализацией и без локализации источников), а также методов нейровизуализации.
ОБСУЖДЕНИЕ
В настоящее время методы математического анализа ЭЭГ, реализованные в различных специализированных программно-аппаратных комплексах, а также в международных свободно распространяемых библиотеках программ, широко используются как в научных исследованиях, так и в медицинской практике. При обработке биоэлектрических сигналов значительное внимание уделяется оценке их пространственной синхронизации, или, иными словами, функциональной коннективности (на основе корреляционного и когерентного анализа, фазовой синхронизации и др. методов). Информативность такого подхода обоснована многолетними фундаментально-научными и клиническими исследованиями (Ливанов, 1972; Гриндель, 1980; Русинов, 1987; Свидерская, 2008; Fries, 2015; Черний и др., 2016; Мачинская и др., 2016; Lee et al., 2017; Изнак и др., 2018; Babiloni et al., 2020). Вместе с тем представление результатов подобных исследований (в цифровом или графическом виде) зачастую носит формальный характер: выводятся все оцениваемые показатели коннективности (фон-реакция, норма-патология и т.п.), удовлетворяющие заданным статистическим критериям. При этом их трактовка, касающаяся оценки соотношения общемозговых и более локальных изменений коннективности, выявления одного или нескольких фокусов активности и степени их взаимозависимости, носит в значительной степени субъективный характер. При расчетах межрегиональных связей получается множество показателей, из которых трудно понять причинно-следственные отношения, выделить совокупности более и менее сгруппированных и соподчиненных церебральных областей (нейросетей).
Анализ литературы, представленной в том числе и в настоящем обзоре, свидетельствует о перспективности ГА, привлекаемого для исследования функциональной активности церебральных областей и связей между ними, для изучения нейросетевой организации головного мозга в норме и при патологии. Теория графов, применяемая в отношении нейронаук, позволяет рассматривать головной мозг как сложную сеть или граф, состоящий из набора вершин и связей между ними. Подобный подход дает возможность выделить основные компоненты сети и рассчитать ряд показателей, описывающих их свойства, а также определить, какие вершины являются центральными (Ушаков, Величковский, 2015; Меринов, 2016; Faber et al., 2020). Для этого могут привлекаться характеристики как анатомических, так и функциональных связей (Friston, 1994; Rubinov, Sporns, 2010; Demertzi et al., 2019; Suárez et al., 2020), основанные на данных методов нейровизуализации (МР-трактография, фМРТ, ПЭТ), электрофизиологии (ЭЭГ, ВП) и МЭГ.
К числу преимуществ ГА относится возможность количественного сопоставления процессов глобальной интеграции (способности быстрого объединения специализированной информации из распределенных областей мозга) и сегрегации (формирования внутри сети сообществ или модулей), происходящих в головном мозге. С помощью данного подхода реально получение информации о функциональной архитектуре сетей мозга, такой как архитектура “малого мира”, а также изучение и моделирование работы нейронных сетей, обусловливающих реализацию определенных когнитивных процессов (как, например, рабочей памяти) (Ioannou, 2014). В табл. 1 настоящего обзора приведен ряд показателей ГА, наиболее часто применяемых в междисциплинарных исследованиях.
Следует отметить, что в электроэнцефалографии вопрос о наличии глобальной и локальной синхронизации биопотенциалов головного мозга и их корково-подкорковой регуляции активно разрабатывался в отечественных научных школах: в экспериментальных исследованиях (Ливанов, 1972), а также в клинических наблюдениях с верифицированной очаговой церебральной патологией (Болдырева и др., 2000). Представления о наличии глобальных и локальных нейронных сетей и их структурно-функциональном обеспечении (Dehaene et al., 2006, 2011) получили широкое распространение с развитием методов нейровизуализации (Meier et al., 2016; Abeyasinghe et al., 2018; Liégeois et al., 2020). Они эффективно используются при исследовании широкого спектра психических и когнитивных процессов (Lin et al., 2014; Lord et al., 2017; Адамович, 2020). ГА рассматривается в качестве адекватного методического подхода и в экспериментальных, и в клинических полидисциплинарных работах данного направления.
К настоящему времени графовый анализ ЭЭГ, применяемый в двух модификациях (без и с локализацией источников генерации биопотенциалов), уже хорошо зарекомендовал себя в различных нейробиологических и клинических задачах. Так, он был успешно использован для поиска ЭЭГ-маркеров болезни Альцгеймера (Zeng et al., 2017; Franciotti et al., 2019; Babiloni et al., 2020; Požar et al., 2020), выявления различий между физиологическими и патологическими признаками старения головного мозга (Knyazev et al., 2015; Miraglia et al., 2016), для обнаружения глобальных и локальных нейросетевых нарушений, характерных для пациентов с фокальной эпилепсией (Hatlestad-Hall et al., 2020), топологических перестроек сети “малого мира” при изменении когнитивных состояний, в онтогенезе (Boersma et al., 2011; Braun et al., 2015; Tóth et al., 2017), при различных неврологических (Fornito et al., 2015) и посттравматических (Farahani et al., 2019) расстройствах, для определения нарушений коннективности ЭЭГ, специфичных по топографии для разных форм патологического угнетения сознания (Cacciola et al., 2019; Carrasco-Gómez et al., 2021). Важно подчеркнуть, что, несмотря на то, что реконструкция нейронных сетей на уровне скальповых отведений ЭЭГ и источников их генерации основывается на отличающихся исходных предположениях, выводы, сделанные по результатам исследований на сходных или идентичных объектах, не противоречат, а лишь дополняют информацию относительно организации функциональных сетей в норме и при разных формах церебральной патологии.
Одним из последних масштабных зарубежных проектов мультимодального анализа коннективности головного мозга с применением графового анализа является Human Connectome Project (Szalkai et al., 2017).
Современные исследователи используют для ГА целый ряд пакетов программ. Некоторые из них представлены в табл. 2 настоящего обзора. При этом освоение пакетов программ Matlab не требует специальных навыков в области программирования. Как правило, для анализа используется линейка toolbox – непосредственно для графового анализа (GraphVar, Brain Connectivity Toolbox и др.) и для визуализации данных (BrainNet Viewer, Connectome Viewer и др.). Для специалистов, обладающих навыками в области программирования, возможно использование пакетов программ на Python (Python NetworkX – для графового анализа, Python MNE – для комплексного анализа нейрофизиологических данных). Во всех вышеперечисленных (табл. 2) toolbox для графового анализа имеется возможность расчета как локальных, так и глобальных метрик сегрегации и интеграции (табл. 1 с метриками).
Математический потенциал графового анализа далеко не исчерпан. Сегодня на практике используется относительно небольшая часть его возможностей. В этой связи есть надежда, что по мере практической адаптации данного подхода будет уточняться информативность уже используемых показателей и расширяться их спектр – для совершенствования не только описания текущего состояния исследуемой системы, но и прогнозирования ее динамики. Кроме того, возможны комбинации ГА с другими методами обработки и визуализации больших объемов данных, в том числе основанными на современных методах машинного обучения, которые могут оказаться перспективными при решении задач прогнозирования.
ЗАКЛЮЧЕНИЕ
Графовый анализ является одним из перспективных методов анализа данных в нейронауках, позволяющим изучать глобальные и локальные характеристики сетей мозга, исследовать особенности передачи информации в нейросетях (поиск активно работающих кластеров, ключевых узлов сети), исследовать динамику изменений коннективности. Данный метод анализа хорошо зарекомендовал себя в работе с различными типами нейрофизиологических данных (ЭЭГ, фМРТ и др.) для решения широкого спектра медицинских задач – диагностических и исследовательских. Современные пакеты программ предоставляют возможность наглядно визуализировать и картировать результаты ГА, представить динамику изменения графовых показателей в виде диаграмм и таблиц, особенности динамической работы нейросетей в виде “графовых карт” головного мозга, что существенно облегчает интерпретацию полученных результатов.
Развитие нейровизуализационных техник в совокупности с новыми методами математической обработки данных ЭЭГ повышает возможности изучения процессов системной нейронной организации, лежащих в основе человеческого сознания, внимания, памяти, принятия решения, а также механизмов их нарушения вследствие травм головного мозга различного генеза. Исследования в данной области в перспективе открывают возможности для разработки новых методов диагностики и лечения.
Список литературы
Адамович Т.В. Интеграция и сегрегация в функциональных сетях головного мозга. Теор. и эксперимент. психол. 2020. 13(3): 1–25.
Анохин К.В. Когнитом: в поисках фундаментальной нейронаучной теории сознания. Журн. высш. нервн. деят. им. И.П. Павлова. 2021. 71(1): 39–71. https://doi.org/10.31857/S0044467721010032
Болдырева Г.Н., Шарова Е.В., Добронравова И.С. Роль регуляторных структур в формировании ЭЭГ человека”. Физ. чел. 2000. 26(5): 19–34.
Гнездицкий В.В. Обратная задача ЭЭГ и клиническая электроэнцефалография. М: Медпрессинформ. 2004. 624 с.
Гриндель О.М. Оптимальный уровень когерентности ЭЭГ и его значение в оценке функционального состояния мозга человека. ЖВНД им. И.П. Павлова. 1980. 30(1): 62–70.
Гриндель О.М., Сазонова О.Б. Введение в клиническую ЭЭГ. Нейрофизиологические исследования в клинике. М.: Антидор. 2001. С. 13–16.
Гутман А.М. Биофизика внеклеточных токов мозга. М: Наука. 1980. 184 с.
Егоров Ю.В., Кузнецова Г.Д. Мозг как объемный проводник. АН СССР. Ин-т высш. нервн. деят. и нейрофиз. М: Наука. 1976. 128 с.
Жаворонкова Л.А. Межполушарная асимметрия мозга человека (правши-левши). 3-е изд., доп. М.: Юрайт. 2019. 217 с.
Иванов Л.Б. Прикладная компьютерная электроэнцефалография. М: МБН. 2004. 352 с.
Изнак А.Ф., Изнак Е.В., Мельникова Т.С. Параметры когерентности ЭЭГ как отражение нейропластичности мозга при психической патологии (обзор литературы). Психиатрия. 2018. 78(2): 127–137. https://doi.org/10.30629/2618-6667-2018-78-127-137
Ливанов М.Н. Пространственная организация процессов головного мозга. М: Наука. 1972. 181 с.
Лисачевa П.Д., Мельников М.Е., Штарк М.Б. Генетические аспекты фМРТ исследований головного мозга. Уcпехи физиол. наук. 2020. 51(1): 58–71. https://doi.org/10.31857/S0301179820010075
Мартынова О.В., Сушинская-Тетерева А.О., Балаев В.В., Иваницкий А.М. Корреляция функциональной связанности областей мозга, активных в состоянии покоя, с поведенческими и психологическими показателями. Журн. высш. нервн. деят. им. И.П.Павлова. 2016. 66(5): 541–555. https://doi.org/10.7868/S0044467716050063
Мачинская Р.И., Розовская Р.И., Курганский А.В., Печенкова Е.В. Корковое функциональное взаимодействие при удержании эмоционально окрашенных изображений в рабочей памяти. Анализ когерентности тета-ритма ЭЭГ в пространстве источников. Физиол. чел. 2016. 42(3): 56–73. https://doi.org/10.7868/S0131164616020120
Меринов А.А., Кочкаров А.А. Анализ сетевой структуры коннектомов человеческого мозга и их классификация. Современ. наука. 2016. 3: 9–16.
Пономарев В.А. Скрытые источники электроэнцефалограммы и связанных с событиями потенциалов и их значение: дис. ИФ им. И.П. Павлова РАН. 2016. 676 с.
Русинов В.С., Гриндель О.М., Болдырева Г.Н., Вакар Е.М. Биопотенциалы мозга человека. М: Медицина. 1987. 254 с.
Русинов В.С. Доминанта. М: Медицина. Акад. мед. наук СССР. 1969. 231 с.
Свидерская Н.Е. Пространственная организация электроэнцефалограммы. М.: Издательство ВГМА. 2008. 573 с.
Ушаков В.Л., Величковский Б.М. Визуализация крупномасштабных сетей головного мозга. Наука и инновации. 2015. 12(154): 22–25.
Фекличева И.В., Чипеева Н.А., Захаров И.М., Исматуллина В.И., Масленникова Е.П., Табуева А.О., Солдатова Е.Л., Малых С.Б. Взаимосвязь интеллекта и функциональной связанности мозга в состоянии покоя. Теор. и эксперимент. психология. 2020. 13(30): 65–80.
Харари Ф. Теория графов. М.: Мир. 1973. 304 с.
Храмов А.Е., Фролов Н.С., Максименко В.А., Куркин С.А., Казанцев В.Б. Функциональные сети головного мозга: от восстановления связей до динамической интеграции. Успехи физических наук. 2021. 191(6): 614–650. https://doi.org/10.3367/UFNr.2020.06.038807
Черний В.И, Андронова И.А., Городник Г.А., Назаренко К.В., Черний Т.В. Исследование ЭЭГ-предикторов нейромедиаторной и нейроглиальной активности, особенностей перестройки межцентральных отношений мозга у пациентов с тяжелой ЧМТ. Межд. невролог. журнал. 2016. 5(83): 41–54. https://doi.org/10.22141/2224-0713.5.83.2016.78470
Шарова Е.В. Асимметрия когерентности ЭЭГ при посткоматозных бессознательных состояниях после тяжелой черепно-мозговой травмы. Функциональная межполушарная асимметрия (хрестоматия) (ред. Боголепова Н.Н., Фокина В.Ф.). М.: Научный мир. 2004. С. 578–585.
Шарова Е.В. Современные возможности ЭЭГ в анализе функциональных нарушений при тяжелых повреждениях головного мозга. Нейронауки: теор. и клин. асп. (Украина). 2009. 5(1–2): 49–58.
Шеповальников А.Н., Цицерошин М.Н., Погосян А.А. О роли различных зон коры и их связей в формировании пространственной упорядоченности поля биопотенциалов мозга в постнатальном онтогенезе. Физиол. чел. 1997. 23(2): 12–24.
Abeyasinghe P.M., Ribeiro de Paula D., Khajehabdollahi S., Valluri S.R., Adrian M., Owen A.M., Soddu A. Role of Dimensionality in Predicting the Spontaneous Behavior of the Brain Using the Classical Ising Model and the Ising Model Implemented on a Structural Connectome. Brain Conect. 2018. 8(7): 444–455. https://doi.org/10.1089/brain.2017.0516
Arnatkeviciūtė A., Fulcher B.D., Pocock R., Fornito A. Hub connectivity, neuronal diversity, and gene expression in the Caenorhabditis elegans connectome. PLoS Comput. Biol. 2018. 14(2): 1–32. https://doi.org/10.1371/journal.pcbi.1005989
Avena-Koenigsberger A., Misic B., Sporns O. Communication dynamics in complex brain networks. Nat. Rev. Neurosci. 2018. 19(1): 17–33.
Awan F.G., Saleem O., Kiran A. Recent trends and advances in solving the inverse problem for EEG source localization. Inverse Probl. Sci. Eng. 2019. 27(11): 1521–1536.
Babiloni C., Blinowska K., Bonanni L., Cichocki A., De Haan W., Claudio del Percio Dubois B., Escudero J., Fernández A., Frisoni G., Guntekin B. Hajos M., Hampel H., Ifeachor E., Kilborn K., Kumar S., Johnsen K., Johannsson M., Jeong J., LeBeau F., Lizio R., Lopes da Silva F., Maestú F., McGeown W.J., McKeith I., Moretti D.V., Nobili F., Olichney J., Onofrj M., Palop J.J., Rowan M., Stocchi F., Struzik Z.M., Tanila H., Teipel S., Taylor J.P., Weiergräber M., Yener G., Young-Pearse T., Drinkenburg W.H., Randall F. What Electrophysiology Tells Us About Alzheimer’s Disease: A Window into the Synchronization and Connectivity of Brain Neurons. Neurobiol. Aging. 2020. 85: 58–73. https://doi.org/10.1016/j.neurobiolaging.2019.09.008
Bassett D.S., Meyer-Lindenberg A., Achard S., Duke T., Bullmore E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc. Natl. Acad. Sci. USA. 2006. 103(51): 19518–19523. https://doi.org/10.1073/pnas.0606005103
Bassett D.S., Sporns O. Network neuroscience. Nat. Neurosci. 2017. 20(3): 353–364.
Bassett D.S., Bullmore E.T. Small-world brain networks revisited. The Neuroscientist. 2017. 23(5): 499–516. https://doi.org/10.1177/1073858416667720
Bernhardt B.C., Bonilha L., Gross D.W. Network analysis for a network disorder: the emerging role of graph theory in the study of epilepsy. Epilepsy & Behavior. 2015. 50: 162–170. https://doi.org/10.1016/j.yebeh.2015.06.005
Boersma M., Smit D.J.A., de Bie H.M.A., Van Baa G.C.M., Boomsma D.I., de Geus E.J.C., Delemarre-van de Waa H.A., Stam C.J. Network analysis of resting state EEG in the developing young brain: structure comes with maturation. Hum. Brain Mapp. 2011. 32(3): 413–425. https://doi.org/10.1002/hbm.21030
Boersma M., Smit D.J.A., Boomsma D.I., De Geus E.J.C., Delemarre-van de Waal H.A., Stam C.J. Growing trees in child brains: graph theoretical analysis of electroencephalography-derived minimum spanning tree in 5-and 7-year-old children reflects brain maturation. Brain connectivity. 2013. 3(1): 50–60. https://doi.org/10.1089/brain.2012.0106
Boldyreva G.N., Zhavoronkova L.A., Sharova E.V., Dobronravova I.S. Electroencephalografic intercentral interaction as a reflection of normal and pathological human brain activity. Span. J. Psychol. 2007. 10(1): 169–179. https://doi.org/10.1017/s1138741600006430
Bordier C., Nicolini C., Bifone A. Graph analysis and modularity of brain functional connectivity networks: searching for the optimal threshold. Front. Neurosci. 2017. 11: 1–9. https://doi.org/10.3389/fnins.2017.0041
Bornholdt S., Schuster H.G. (eds.) Handbook of graphs and networks: from the genome to the Internet. Weinheim: Wiley-VCH. 2003. 416 p. https://doi.org/10.1002/3527602755
Bosch-Bayard J., Girini K., Biscay R.J., Valdes-Sosa P., Evans A.C., Chiarenza G.A. Resting EEG effective connectivity at the sources in developmental dysphonetic dyslexia. Differences with non-specific reading delay. Int. J. Psychophysiol. 2020. 153: 135–147. https://doi.org/10.1016/j.ijpsycho.2020.04.021
Buchanan C.R., Bastin M.E., Ritchie S.J., Liewald D.C., Madole J.W., Tucker-Drob E.M., Deary I.J., Cox S.R. The effect of network thresholding and weighting on structural brain networks in the UK Biobank. NeuroImage. 2020. 211: 1–14. https://doi.org/10.1016/j.neuroimage.2019.116443
Buckner R.L., Sepulcre J., Talukdar T., Krienen F.M., Liu H., Hedden T., Johnson K.A. Cortical Hubs Revealed by Intrinsic Functional Connectivity: Mapping, Assessment of Stability, and Relation to Alzheimer’s Disease. J. Neurosci. 2009. 29(6): 1860–1873. https://doi.org/10.1523/jneurosci.5062-08.2009
Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009. 10(3): 186–198. https://doi.org/10.1038/nrn2575
Bullmore E., Sporns O. The economy of brain network organization. Nat. Rev. Neurosci. 2012. 13(5): 336–349. https://doi.org/10.1038/nrn3214
Cacciola A., Naro A., Milardi D., Bramanti A., Malatacca L., Spitaleri M., Leo A., Muscoloni A., Cannistraci C.V., Bramanti P., Calabrò R.S., Anastasi G.P. Functional brain network topology discriminates between patients with minimally conscious state and unresponsive wakefulness syndrome. Int. J. Clin. Med. 2019. 8(3): 1–22. https://doi.org/10.3390/jcm8030306
Cao Z., Lin C.T., Chuang C.H., Lai K.L., Yang A.C., Fuh J.L., Wang S.J. Resting-state EEG power and coherence vary between migraine phases. J. Headache Pain. 2016. 17(1): 1–9. https://doi.org/10.1186/s10194-016-0697-7
Cao R., Hao Y., Wang X., Gao Y., Shi H., Huo S., Wang B., Guo H., Xiang J. EEG Functional Connectivity Underlying Emotional Valance and Arousal Using Minimum Spanning Trees. Front. Neurosci. 2020. 14: 1–12. https://doi.org/10.3389/FNINS.2020.00355/BIBTEX
Carrasco-Gómez M., Keijzer H.M., Ruijter B.J., Bruña R., Tjepkema-Cloostermans M.C., Hofmeijer J., van Putten M.J. EEG functional connectivity contributes to outcome prediction of postanoxic coma. Clin Neurophysiol. 2021. 132: 1312–1320. https://doi.org/10.1016/j.clinph.2021.02.011
Chen Z.J., He Y., Rosa-Neto P., Germann J., Evansa C. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb. Cortex. 2008. 18(10): 2374–2381. https://doi.org/10.1093/cercor/bhn003
Chen Y., Huang X., Wu M., Li K., Hu X., Jiang P., Chen L., He N., Dai J., Wang S., He M., Guo L., Sweeney J.A., Gong Q. Disrupted brain functional networks in drug-naive children with attention deficit hyperactivity disorder assessed using graph theory analysis. Hum Brain Mapp. 2019. 40(17): 4877–4887. https://doi.org/10.1002/hbm.24743
Chennu S., Finoia P., Kamau E., Allanson J., Williams G.B., Monti M.M., Noreika V., Arnatkeviciute A., Canales-Johnson A., Olivares F., Cabezas-Soto D., Menon D.K., Pickard J.D., Owen A.M., Bekinschtein T.A. Spectral Signatures of Reorganised Brain Networks in Disorders of Consciousness. PLoS Comput Biol. 2014. 10(10): 1–16. https://doi.org/10.1371/journal.pcbi.1003887
Chennu S., Annen J., Wannez S., Thibaut A., Chatelle C., Cassol H., Martens G., Schnakers C., Gosseries O., Menon D., Laureys S. Brain networks predict metabolism, diagnosis and prognosis at the bedside in disorders of consciousness. Brain. 2017. 140(8): 2120–2132. https://doi.org/10.1093/brain/awx163
Collin G., van den Heuvel M.P. The ontogeny of the human connectome: development and dynamic changes of brain connectivity across the life span. Neuroscientist. 2013. 19(6): 616–628. https://doi.org/10.1177/1073858413503712
Crobe A., Demuru M., Didaci L., Marcialis G.L., Fraschini M. Minimum spanning tree and k-core decomposition as measure of subject-specific EEG traits. Biomed. Phys. Eng. Express 2016. 2(1): 1–16. https://doi.org/10.1088/2057-1976/2/1/017001
Dai Z., de Souza J., Lim J., Ho P.M., Chen Y., Li J., Thakor N., Bezerianos A., Sun Y. EEG cortical connectivity analysis of working memory reveals topological reorganization in theta and alpha bands. Front. Hum. Neurosci. 2017. 11: 1–13. https://doi.org/10.3389/fnhum.2017.00237
Dai Z., Lin Q., Li T., Wang X., Yuan H., Yu X., He Y., Wang H. Disrupted structural and functional brain networks in Alzheimer’s disease. Neurobiol. Aging. 2019. 75: 71–82. https://doi.org/10.1016/j.neurobiolaging.2018.11.005
Dehaene S., Changeux J.-P., Naccache L., Sackur J., Sergent C. Conscious, preconscious, and subliminal processing: a testable taxonomy. Trends Cogn. 2006. 10(5): 204–211. https://doi.org/10.1016/j.tics.2006.03.007
Dehaene S., Changeux J.-P., Naccache L. The Global Neuronal Workspace Model of Conscious Access: From Neuronal Architectures to Clinical Applications. In: Dehaene S., Christen Y. (eds) Characterizing Consciousness: From Cognition to the Clinic? Research and Perspectives in Neurosciences. Springer, Berlin, Heidelberg. 2011. 18: 55–84. https://doi.org/10.1007/978-3-642-18015-6_4
Dell’Italia J., Johnson M.A., Vespa P.M., Monti M.M. Network analysis in disorders of consciousness: four problems and one proposed solution (exponential random graph models). Front. Neurol. 2018. 9: 439–459.
Demertzi A., Tagliazucchi E., Dehaene S., Deco G., Barttfeld P., Raimondo F., Martial C., Fernández-Espejo D., Rohaut B., Voss H.U., Schiff N.D., Owen A.M., Laureys S., Naccache L., Sitt J.D. Human consciousness is supported by dynamic complex patterns of brain signal coordination. Sci. Adv. 2019. 5(2): 1–11. https://doi.org/10.1126/sciadv.aat7603
Dickie D.A., Shenkin S.D., Anblagan D., Lee J., Blesa C.M., Rodriguez D., Wardlaw J.M. Whole Brain Magnetic Resonance Image Atlases: A Systematic Review of Existing Atlases and Caveats for Use in Population Imaging. Front. Neuroinform. 2017. 11: 1–15. https://doi.org/10.3389/fninf.2017.00001
Douw L., Schoonheim M.M., Landi D., Van der Meer M.L., Geurts J.J., Reuneveld J.C., Klein M., Stam C.J. Cognition is related to resting-state small-world network topology: an magnetoencephalographic study. Neuroscience. 2011. 175: 169–177. https://doi.org/10.1016/j.neuroscience.2010.11.039
Douw L., van Dellen E., Gouw A.A., Griffa A., de Haan W., van den Heuvenl M., Hillebrand A., van Meighem P., Nissen I.A., Otte W.M., Reijmer Y.D., Schoonheim M.M., Senden M., van Straaten E.C.W., Tijms B.M., Tewarie P., Stam C.J. The road ahead in clinical network neuroscience. Network Neuroscience. 2019. 3(4): 969–993. https://doi.org/10.1162/netn_a_00103
Faber J., Antoneli P.C., Via G., Araújo N.S., Pinheiro D.J.L.L., Cavalheiro E. Critical elements for connectivity analysis of brain networks. In: Tsytsarev V., Yamamoto V., Zhong N. (eds) Functional Brain Mapping: Methods and Aims. 2020. P. 67–107. Brain Inform. https://doi.org/10.1007/978-981-15-6883-1_4
Fagiolo G. Clustering in complex directed networks. Phys. Rev. E. 2007. 76(2): 1–16.
Farahani F.V., Karwowski W., Lighthall N.R. Application of graph theory for identifying connectivity patterns in human brain networks: a systematic review. Frontiers Neurosci. 2019. 13: 1–27. https://doi.org/10.3389/fnins.2019.00585
Farashi S., Khosrowabadi R. EEG based emotion recognition using minimum spanning tree. Phys. Eng. Sci. Med. 2020. 43(3): 985–996. https://doi.org/10.1007/s13246-020-00895-y
Finotelli P., Forlim C.G., Klock L., Pini A., Bächle J., Stoll L., Giemsa P., Fuchsd M., Schoofs N., Montag C., Dulioa P., Gallinat J., Kühnb S. New graph-theoretical-multimodal approach using temporal and structural correlations reveals disruption in the thalamo-cortical network in patients with schizophrenia. Brain Connect. 2019. 9(10): 760–769. https://doi.org/10.1089/brain.2018.0654
Fornito A., Bullmore E.T. Connectomics: a new paradigm for understanding brain disease. Eur. Neuropsychopharmacol. 2015. 25(5): 733–748. https://doi.org/10.1016/j.euroneuro.2014.02.011
Fox P.T., Friston K.J. Distributed processing: distributed functions? Neuroimage. 2012. 61(2): 407–426. https://doi.org/10.1016/j.neuroimage.2011.12.051
Franciotti R., Falasca N.W., Arnaldi D., Famà F., Babiloni C., Onofrj M., Nobili F.M., Bonanni L. Cortical network topology in prodromal and mild dementia due to Alzheimer’s disease: graph theory applied to resting state EEG. Brain topogr. 2019. 32(1): 127–141. https://doi.org/10.1007/s10548-018-0674-3
Fries P. Rhythms for Cognition: Communication Through Coherence. Neuron. 2015. 88(1): 220–235. https://doi.org/10.1016/j.neuron.2015.09.034
Friston K.J., Frith C.D., Liddle P.F., Frackowiak R.S. Functional connectivity: the principal- component analysis of large (PET) data sets. J. Cereb. Blood Flow Metab. 1993. 13(1): 5–14. https://doi.org/10.1038/jcbfm.1993.4
Friston K.J. Functional and effective connectivity in neuroimaging: a synthesis. Hum. brain mapp. 1994. 2(1–2): 56–78.
Friston K.J. The free-energy principle: a rough guide to the brain? Trends Cogn. 2009. 13(7): 293–301. https://doi.org/10.1016/j.tics.2009.04.005
Girn M., Mills C., Christoff K. Linking brain network reconfiguration and intelligence: Are we there yet? Trends Neurosci. Educ. 2019. 15: 62–70. https://doi.org/10.1016/j.tine.2019.04.001
Gong G., He Y., Concha L., Lebel C., Gross D.W., Evans A.C., Beaulieu C. Mapping Anatomical Connectivity Patterns of Human Cerebral Cortex Using In Vivo Diffusion Tensor Imaging Tractography.Cereb. Cortex. 2009. 19(3): 524–536. https://doi.org/10.1093/cercor/bhn102
Guye M., Bettus G., Bartolomei F., Cozzone P.J. Graph theoretical analysis of structural and functional connectivity MRI in normal and pathological brain networks. MAGMA. 2010. 23(5): 409–421. https://doi.org/10.1007/s10334-010-0205-z
Habas Ch. (Ed). The Neuroimaging of Brain Diseases. Springer Int. Publishing AG. 2018. 353 p. https://doi.org/10.1007/978-3-319-78926-2
Hagmann P., Kurant M., Gigandet X., Thiran P., Wedeen V.J., Meuli R., Thiran J.P. Mapping human whole-brain structural networks with diffusion MRI. PloS one. 2007. 2(7): 1–9. https://doi.org/10.1371/journal.pone.0000597
Hagmann P., Cammoun L., Gigandet X., Meuli R., Honey C.J., Wedeen V.J., Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008. 6(7): 1479–1493. https://doi.org/10.1371/journal.pbio.0060159
Hamzeh M., Vahidi B., Nematollahi A.F. Optimizing configuration of cyber network considering graph theory structure and teaching learning-based optimization (GT-TLBO). IEEE Trans. Industr. Inform. 2019. 15(4): 2083–2090. https://doi.org/10.1109/TII.2018.2860984
Hassan M., Dufor O., Merlet I., Berrou C., Wendling F. EEG source connectivity analysis: from dense array recordings to brain networks. PloS one. 2014. 9(8): 1–15. https://doi.org/10.1371/journal.pone.0105041
Hasan M.A.H., Khan M.U., Mishra D.A. Computationally Efficient Method for Hybrid EEG-fNIRS BCI Based on the Pearson Correlation. BioMed Research International. 2020. 2020: 1–13. https://doi.org/10.1155/2020/1838140
Hatlestad-Hall C., Bruña R., Syvertsen M.R., Erichsen A., Andersson V., Vecchio F., Miraglia F., Rossini P. M., Renvall H., Taubøll E., Maestú F., Haraldsen I.H. Source-level EEG and graph theory reveal widespread functional network alterations in focal epilepsy. Clinical Neurophysiology. 2021. 132(7): 1663–1676.
He Y., Chen Z.J., Evans A.C. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb. Cortex. 2007. 17(10): 2407–2419. https://doi.org/10.1093/cercor/bhl149
Hilgetag C.C., Kaiser M. Clustered organization of cortical connectivity. Neuroinformatics. 2004. 2(2): 353–360. https://doi.org/10.1385/NI:2:3:353
Hilgetag C.C., Beul S.F., van Albada S.J., Goulas A. An architectonic type principle integrates macroscopic cortico-cortical connections with intrinsic cortical circuits of the primate brain. Network neuroscience. 2019. 3(4): 905–923. https://doi.org/10.1162/netn_a_00100
Humphries M.D., Gurney K. Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PloS one. 2008. 3(4): 1–10. https://doi.org/10.1371/journal.pone.0002051
Ioannou P. Computational Modelling of Working Memory. PhD thesis. University of Surrey (United Kingdom). 2014. P. 1–104.
Ion A., Kropatsch W.G., Haxhimusa Y. Considerations regarding the minimum spanning tree pyramid segmentation method. In: Proc. 11 th Workshop Structural, Syntactic, and Statistical Pattern Recognition (SSPR). LNCS. 2006. 4109: 182–190.
Iturria-Medina Y., Sotero R.C., Canales-Rodriguez E.J., Aleman-Gomez Y., Melie-Garcia L. Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. Neuroimage. 2008. 40(3): 1064–1076.
Jatoi M.A., Kamel N. Brain source localization using EEG signal analysis. CRC Press. 2017. 248 p.
Kennedy H., Van Essen D.C., Christen Y. Micro-, Meso- and Macro-Connectomics of the Brain Springer Nature. 2016. 166 p.
Knyazev G.G., Volf N.V., Belousova L.V. Age-related differences in electroencephalogram connectivity and network topology.Neurobiology of aging. 2015. 36(5): 1849–1859.
Koessler L., Maillard L., Benhadid A., Vignal J.P., Felblinger J., Vespignani H., Braun M. Automated cortical projection of EEG sensors: anatomical correlation via the international 10–10 system. Neuroimage. 2009. 46(1): 64–72.
Kunszenti-Kovács D., Lovász L., Szegedy B. Measures on the square as sparse graph limits. J. Comb. Theory. Ser. B. 2019. 138: 1–40.
Larsen J.C. Electrical network theory of countable graphs. IEEETrans. Circuits Syst. I. Regul. Pap. 1997. 44(11): 1045–1055.
Latora V., Marchiori M. Efficient behavior of small-world networks. Phys. Rev. Lett. 2001. 87: 1–4. https://doi.org/10.1103/PhysRevLett.87.198701
Liao X., Vasilakos A.V., He Y. Small-world human brain networks: perspectives and challenges. Neurosci. Biobehav. Rev. 2017. 77: 286–300. https://doi.org/10.1016/j.neubiorev.2017.03.018
Liégeois R., Ziegler E., Geurts P., Gomez F., Bahri M.A., Phillips C., Soddu A., Vanhaudenhuyse A., Laureys S., Sepulchre R. Cerebral functional connectivity periodically (de)synchronizes with anatomical constraints. Brain Struct. Func. 2014. 221(6): 2985–2997. https://doi.org/10.1007/s00429-015-1083-y
Liégeois R., Santos A., Matta V., Van De Ville D., Sayed A.H. Revisiting correlation-based functional connectivity and its relationship with structural connectivity. Network Neurosci. 2020. 4(4): 1235–1251.
Lin P., Sun J., Yu G., Wu Y., Yang Y., Liang M., Liu X. Global and local brain network reorganization in attention-deficit/hyperactivity disorder. Brain Imaging Behav. 2014. 8(4): 558–569. https://doi.org/10.1007/s11682-013-9279-3
Lopes da Silva F. EEG: Origin and Measurement. EEG-fMRI, Physiological Basis, Technique and Applications. Mulert C., Lemieux L. (eds.) Berlin Heidelberg: Springer Verlag. 2010. 968 p.
Lord L.D., Stevner A.B., Deco G., Kringelbach M.L. Understanding principles of integration and segregation using whole-brain computational connectomics: implications for neuropsychiatric disorders. Philos. Trans. A. Math. Phys. Eng. Sci. 2017. 375: 1–21. https://doi.org/10.1098/rsta.2016.0283
Maksimenko V.A., Runnova A.E., Frolov N.S., Makarov V.V., Nedaivozov V., Koronovskii A.A., Pisarchik A., Alexander E. Hramov A.E. Multiscale neural connectivity during human sensory processing in the brain. Phys. Rev. E. 2018. 97: 1–10. https://doi.org/10.1103/PhysRevE.97.052405
Mannil M., Winklhofer S.F.-X. (ed). Neuroimaging Techniques in Clinical Practice. Springer Nature Switzerland AG. 2020. 342 p. https://doi.org/10.1007/978-3-030-48419-4
McColgan P., Blom T., Rees G., Seunarine K.K., Gregory S., Johnson E., Durr A., Roos R., Scahill1 R.I., Clark C.A., Tabrizi S.J., Adeel Razi A. Stability and sensitivity of structural connectomes: effect of thresholding and filtering and demonstration in neurodegeneration. bioRxiv preprint. https://doi.org/10.1101/416826
Meier J., Tewarie P., Hillebrand A., Douw L., van Dijk B.W., Stufflebeam S.M., Van Mieghem P. Mapping Between Structural and Functional Brain Networks. Brain Connectivity. 2016. 6(4): 298–311. https://doi.org/10.1089/brain.2015.0408
Michel C.M., Brunet D. EEG source imaging: a practical review of the analysis steps. Frontiers in neurology. 2019. 10: 1–18. https://doi.org/10.3389/fneur.2019.00325
Micheloyannis S. Graph-based network analysis in schizophrenia.World J Psychiatry. 2012. 2(1): 1–12. https://doi.org/10.5498/wjp.v2.i1.1
Micheloyannis S., Pachou E., Stam C.J., Breakspear M., Bitsios P., Vourkas M., Zervakis M. Small-world networks and disturbed functional connectivity in schizophrenia.Schizophrenia research. 2006. 87(1–3): 60–66.
Miraglia F., Vecchio F., Bramanti P., Rossini P.M. EEG characteristics in “eyes-open” versus “eyes-closed” conditions: small-world network architecture in healthy aging and age-related brain degeneration. Clinical Neurophysiology. 2016. 127(2): 1261–1268.
Newman M.E.J. The Structure and Function of Complex Networks. SIAM Review. 2003. 45(2): 167–256.
Newman M.E.J. Modularity and community structure in networks.Proceedings of the National Academy of Sciences. United States National Academy of Sciences. 2006. 103(23): 8577–8696. https://doi.org/10.1073/pnas.0601602103
Nunez P.L. Electric fields of the brain. The Neurophysics of EEG. Oxford University Press. 1981. 626 p.
Olejarczyk E., Jernajczyk W. Graph-based analysis of brain connectivity in schizophrenia. PLoS One. 2017. 12(11): 1–28. https://doi.org/10.1371/journal.pone.0188629
Onnela J.P., Saramäki J., Kertész J., Kaski K. Intensity and coherence of motifs in weighted complex networks. Physical Review E. 2005. 71(6): 1–5. https://doi.org/10.1103/physreve.71.065103
Pascual-Marqui R.D., Biscay R.J., Bosch-Bayard J., Lehmann D., Kochi K., Kinoshita T., Yamada N., Sadato N. Assessing direct paths of intracortical causal information flow of oscillatory activity with the isolated effective coherence (iCoh). Front. Hum. Neurosci. 2014. 8(448): 1–12. https://doi.org/10.3389/fnhum.2014.00448
Pascual-Marqui R.D., Esslen M., Kochi K., Lehmann D. Functional imaging with low-resolution brain electromagnetic tomography (LORETA): a review. Methods Find Exp Clin Pharmacol. 2002. 24: 91–95.
Pascual-Marqui R.D., Lehmann D., Koukkou M., Kochi K., Anderer P., Saletu B., Tanaka H., Hirata K., John E.R., Prichep L., Biscay-Lirio R., Kinoshita T. Assessing interactions in the brain with exact low-resolution electromagnetic tomography. Philos. Trans. R. Soc. A. 2011. 369(1952): 3768–3784.
Peraza L.R., Asghar A.U., Green G., Halliday D.M. Volume conduction effects in brain network inference from electroencephalographic recordings using phase lag index. J Neurosci Methods. 2012. 207(2): 189–199. https://doi.org/10.1016/j.jneumeth.2012.04.007
Pfurtscheller G., Schwerdtfeger A.R., Seither-Preisler A, Brunner C., Stefan A.C., Brito J., Carmo M.P., Andrade A. Brain-heart communication: Evidence for “central pacemaker” oscillations with a dominant frequency at 0.1 Hz in the cingulum. Clin. Neurophysiol. 2017. 128(1): 183–193. https://doi.org/10.1016/j.clinph.2016.10.097
Phillips D.J., McGlaughlina A., Rutha D., Jager L.R., Soldan A. Graph theoretic analysis of structural connectivity across the spectrum of Alzheimer’s disease: The importance of graph creation methods. NeuroImage: Clin. 2015. 2015(7): 377–390. https://doi.org/10.1016/j.nicl.2015.01.007
Pospelov N., Nechaev S., Anokhin K., Valba O., Avetisov V., Gorsky A. Spectral peculiarity and criticality of a human connectome. Phys. Life. Rev. 2021. 31: 240–256.
Požar R., Giordani B., Kavcic V. Effective differentiation of mild cognitive impairment by functional brain graph analysis and computerized testing. PloS one. 2020. 15(3): 1–19. https://doi.org/10.1371/journal.pone.0230099
Proshina E.A., Savostyanov A.N., Bocharov A.V., Knyazev G.G. Effect of 5-HTTLPR on current source density, connectivity, and topological properties of resting state EEG networks. Brain research, 2018. 1697: 67–75. https://doi.org/10.1016/j.brainres.2018.06.018
Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations.Neuroimage. 2010. 52(3):1059–1069
Rubinov M., Ypma R.J., Watson C., Bullmore E.T. Wiring cost and topological participation of the mouse brain connectome.Proc. Natl. Acad. Sci. USA. 2015. 112(32): 10032–10037. https://doi.org/10.1073/pnas.1420315112
Saramäki J., Kivelä M., Onnela J.P., Kaski K., Kertész J. Generalizations of the clustering coefficient to weighted complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2007. 75(2): 1–4. https://doi.org/10.1103/PhysRevE.75.027105
Schoffelen J.M., Gross J. Source connectivity analysis with MEG and EEG.Hum Brain Mapp. 2009. 30: 1857–1865. https://doi.org/10.1002/hbm.20745
Sharaev M.G., Zavyalova V.V., Ushakov V.L., Kartashov S.I., Velichkovsky B.M. Effective Connectivity within the Default Mode Network: Dynamic Causal Modeling of Resting-State fMRI Data.Front. Hum. Neurosci. 2016. 10: 1–9. https://doi.org/10.3389/fnhum.2016.00014
Sharova E.V., Pogosbekyan E.L., Korobkova E.V., Zaitsev O.S., Zakharova N.E., Chelyapina M.V., Fadeeva L.M., Oknina L.B., Pronin I.N., Potapov A.A. Interhemispheric connectivity and attention in patients with disorders of consciousness after severe traumatic brain injury. J. Neurol. Stroke. 2018. 8(4): 245‒253. https://doi.org/10.15406/jnsk.2018.08.00319
Shine J.M., Breakspear M., Bell P.T., Ehgoetz Martens K.A., Shine R., Koyejo O., Sporns O., Poldrack R.A. Human cognition involves the dynamic integration of neural activity and neuromodulatory systems. Nat. Neurosci. 2019. 22(2): 289–296. https://doi.org/10.1038/s41593-018-0312-0
Simard D., Nadeau L., Kröger H. Fastest learning in small-world neural networks. Phys. Lett. A. 2005. 336(1): 8–15.
Singer W. Distributed processing and temporal codes in neuronal networks.Cogn. Neurodyn. 2009. 3(3): 189–196.
Sporns O., Zwi J.D. The small world of the cerebral cortex. Neuroinformatics. 2004. 2(2): 145–162.
Sporns O., Honey C.J., Kötter R. Identification and classification of hubs in brain networks. PloS one. 2007. 2(10): 10–49.
Sporns O. Networks of the Brain. Cambridge, MA: MIT Press. 2010. 424 p.
Sporns O. Graph theory methods: applications in brain networks. Dialogues in clinical neuroscience. 2018. 20(2): 111–121.
Steriade M., Gloor P., Llinás R.R., Lopes da Silva F.H., Mesulam M.M. Basic mechanisms of cerebral rhythmic activities. EEG and Clinical Neurophysiology. 1990. 76(6): 481–508.
Stumme J., Jockwitz C., Hoffstaedter F., Amunts K., Caspers S. Functional network reorganization in older adults: Graph-theoretical analyses of age, cognition and sex. NeuroImage. 2020. 214: 1–15. https://doi.org/10.1016/j.neuroimage.2020.116756
Suárez L.E., Markello R.D., Betzel R.F., Misic B. Linking structure and function in macroscale brain networks.Tr. Cog. Sci. 2020. 24(4): 302–315. https://doi.org/10.1016/j.tics.2020.01.008
Szalkai B., Kerepesi C., Varga B., Grolmusz V. Parameterizable consensus connectomes from the human connectome project: The budapest reference connectome server v3. 0. Cogn. neurodyn. 2017. 11(1): 113–116. https://doi.org/10.1007/s11571-016-9407-z
Tan B., Kong X., Yang P., Jin Z., Li L. The difference of brain functional connectivity between eyes-closed and eyes-open using graph theoretical analysis. Comput Math Methods Med. 2013. 2013: 1–15. https://doi.org/10.1155/2013/976365
Termenon M., Jaillard A., Delon-Martin C., Achard S. Reliability of graph analysis of resting state fMRI using test-retest dataset from the Human Connectome Project. Neuroimage. 2016. 142: 172–187. https://doi.org/10.1016/j.neuroimage.2016.05.062
Tononi G., Sporns O., Edelman G.M. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proceedings of the National Academy of Sciences. 1994. 91(11): 5033–5037.
Tóth B., Urbán G., Háden G.P., Márk M., Török M., Stam C.J., Winkler I. Large-scale network organization of EEG functional connectivity in newborn infants.Human brain mapping. 2017. 38(8): 4019–4033. https://doi.org/10.1002/hbm.23645
Ustimenko V. On computations with Double Schubert Automaton and stable maps of Multivariate Cryptography. arXiv preprint. 2021. https://doi.org/10.48550/arXiv.2108.08288
Vaessen M.J., Hofman P.A., Tijssen H.N., Aldenkamp A.P., Jansen J.F., Backes W.H. The effect and reproducibility of different clinical DTI gradient sets on small world brain con-nectivity measures. Neuroimage. 2010. 51(3): 1106–1116.
Van den Heuvel M.P., Hulshoff Pol H.E. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 2010. 20: 519–534. https://doi.org/10.1016/j.euroneuro.2010.03.008
Van den Heuvel M.P., Sporns O. An Anatomical Substrate for Integration among Functional Networks in Human Cortex. J. Neurosci. 2013. 33(36): 14489–14500. https://doi.org/10.1523/jneurosci.2128-13.2013
Van den Heuvel M.P., Sporns O. A cross-disorder connectome landscape of brain dysconnectivity. Nat. rev. neurosci. 2019. 20(7): 435–446.
Vecchio F., Miraglia F., Rossini M.P. Connectome: Graph theory application in functional brain network architecture. Clin. Neurophysiol. Pract. 2017. 24(2): 206–213. https://doi.org/10.1016/j.cnp.2017.09.003
Vecchio F., Gorgoni M., Ferrara M., Iberite F., Bramanti P., Gennaro L., Rossini P.M. Cortical connectivity modulation during sleep onset: A study via graph theory on EEG data. Hum. Brain Mapp. 2017. 38(11): 5456–5464. https://doi.org/10.1002/hbm.23736
Vecchio F., Miraglia F., Rossini P.M. Connectome: Graph theory application in functional brain network architecture. Clinical Neurophysiology. 2017. 2: 206–213. https://doi.org/10.1016/j.cnp.2017.09.003
Viola E., Wigderson A. Local expanders. Comput. Complex. 2018. 27(2): 225–244.
Wang J., Zuo X., He Y. Graph-based network analysis of resting-state functional MRI. Front. Syst. Neurosci. 2010. 4(16): https://doi.org/10.3389/fnsys.2010.00016
Watanabe H., Shinoda S. Soul of circuit theory. a review on research activities of graphs and circuits in Japan. IEEE Transactions on Circuits and Systems I: Fundamental Theory and Applications. 1999. 46(1): 83–94.
Watts D.J., Strogatz S.H. Collective dynamics of ‘small-world’networks. Nature. 1998. 393 (6684): 440–442.
Wendelken C., Ferrer E., Ghetti S., Bailey S.K., Cutting L., Bunge S.A. Frontoparietal structural connectivity in childhood predicts development of functional connectivity and reasoning ability: A large-scale longitudinal investigation. J. Neurosci. 2017. 37(35): 8549–8558. https://doi.org/10.1523/JNEUROSCI.3726-16.2017
Wu G., Munsell B., Laurienti P., Chung M.K. GRAND: Unbiased Connectome Atlas of Brain Network by Groupwise Graph Shrinkage and Network Diffusion. In: Wu G., Rekik I., Schirmer M., Chung A., Munsell B. (eds) Connectomics in NeuroImaging. CNI 2018. Lecture Notes in Computer Science. Springer, Cham. 2018. 11083: 127–135. https://doi.org/10.1007/978-3-030-00755-3_14
Xia M., Wang J., He Y. BrainNet Viewer: a network visualization tool for human brain connectomics. PloS one. 2013. 8(7): 1–15. https://doi.org/10.1371/journal.pone.0068910
Zeng K., Kang J., Ouyang G., Li J., Han J., Wang Y., Sokhadze E.M., Casanova M.F., Li X. Disrupted Brain Network in Children with Autism Spectrum Disorder. Sci. Rep. 2017. 7(1): 1–12. https://doi.org/10.1038/s41598-017-16440-z
Zhu G., Li Y., Wen P.P. Analysis and Classification of Sleep Stages Based on Difference Visibility Graphs From a Single-Channel EEG Signal. IEEE Journal of Biomedical and Health Informatics. 2014. 18(6): 1813–1821. https://doi.org/10.1109/JBHI.2014.2303991
Дополнительные материалы отсутствуют.
Инструменты
Журнал высшей нервной деятельности им. И.П. Павлова


